Figure 6.
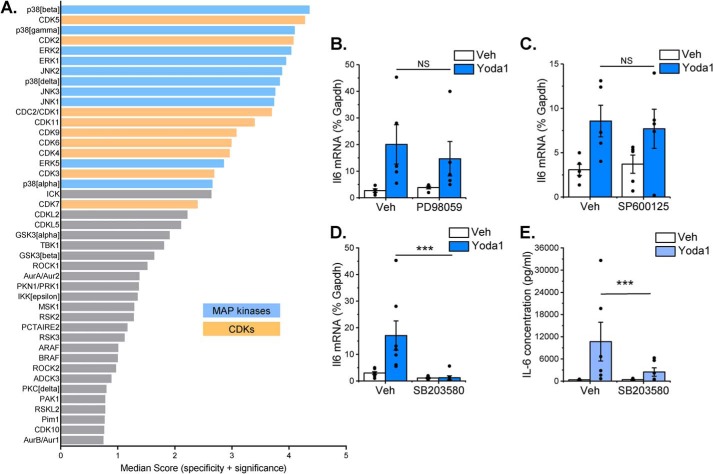
Piezo1 activation induces MAP kinase signaling, with p38 MAPK being coupled to IL-6 expression. A, mouse cardiac fibroblasts were stimulated with 10 μm Yoda1 for 10 min before analyzing Ser/Thr protein kinase activity with PamChip multiplex kinase activity profiling (n = 6). Kinases were identified based on peptide substrate phosphorylation and ranked by specificity and statistical significance (only the top 40 are shown). See Table S1 for the full dataset. The top kinase families predicted to be activated by Yoda1 were the MAPKs ERK1/2/5, JNK1/2/3, and p38α/β/γ/δ (blue) and CDKs 1–7, 9, and 11 (orange). B–D, RT-PCR analysis of Il6 mRNA expression after exposure of murine cardiac fibroblasts to 30 μm PD98059 (B, an ERK pathway inhibitor), 10 μm SP600125 (C, a JNK inhibitor), or 10 μm SB203580 (D, a p38 MAPK inhibitor) for 1 h, followed by treatment with vehicle (Veh) or 10 μm Yoda1 for a further 24 h. Expression is measured as percent of the housekeeping control Gapdh (n = 7 for SB203580, n = 5 for PD98059, and SP600125). Repeated measures two-way ANOVA for B: p = 0.7738, F = 0.133 (PD98059); p = 0.0529, F = 7.41 (Yoda1); p = 0.069, F = 6.08 (interaction). For C: p = 0.6133, F = 0.299 (SP600125); p = 0.0020, F = 51.9 (Yoda1); p = 0.475, F = 0.62 (interaction). For D: p = 0.0004, F = 52.1 (SB203580); p = 0.2942, F = 1.32 (Yoda1); p = 0.0028, F = 23.8 (interaction). Post hoc test: ***, p < 0.001; NS, not significant for the effect of inhibitor. E, murine cardiac fibroblasts were treated with vehicle or 10 μm SB203580 for 1 h and then treated with either vehicle or 10 μm Yoda1 for 24 h before collecting conditioned media and measuring IL-6 levels by ELISA (n = 6). Repeated measures two-way ANOVA: p = 0.0217, F = 10.8 (SB203580); p = 0.0283, F = 9.3 (Yoda1); p = 0.0002, F = 98.1 (interaction). Post hoc test: ***, p < 0.001 for the effect of SB203580.