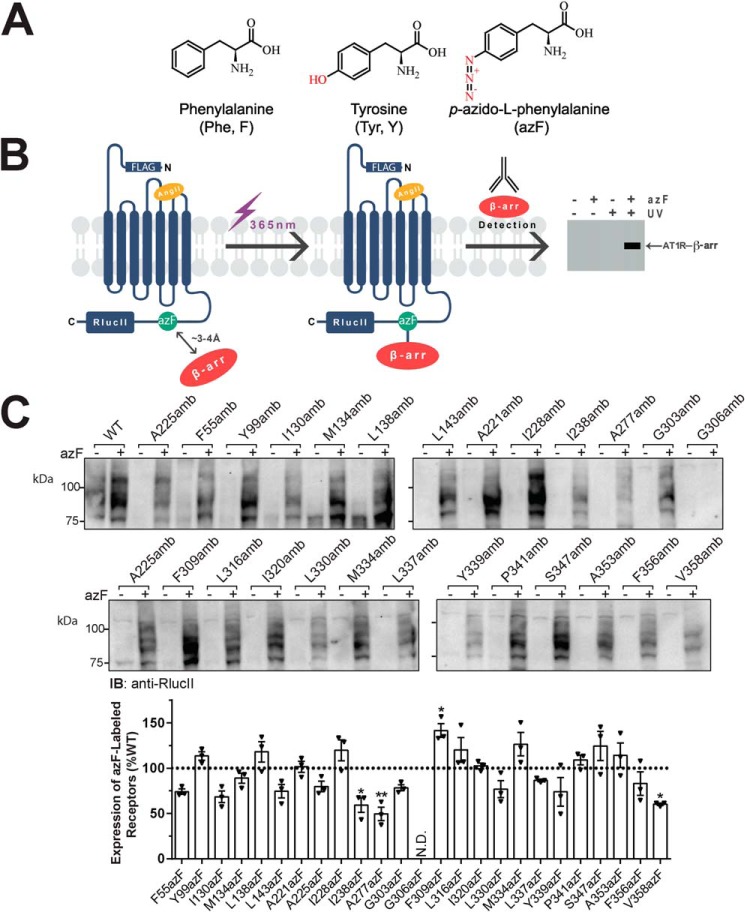
Figure 1.
Chemical structures of natural and unnatural amino acids, photocross-linking approach, and azF-incorporated AT1R mutant expression. A, chemical structures of phenylalanine and tyrosine with three-letter and single-letter nomenclature and azF resonance structure (two double bonds). B, schematic of the photocross-linking approach. All experiments were conducted on AT1R containing an N-terminal FLAG tag and a C-terminal RlucII epitope. AzF is incorporated at site-specific amber (TAG) mutations using a heterologous cell system and the amber codon suppression technology as described under “Experimental procedures.” Cells expressing azF-incorporated AT1Rs along with β-arr1 are incubated with AngII followed by exposure to UV light (365 nm). Photoactivation of azF in AT1R promotes the formation of a covalent bond with primary amines or aliphatic hydrogens lying within its proximity (up to 3–4 Å), allowing capturing β-arr through cross-linking. The AT1R–β-arr complex is then immunologically detected to reveal its size. C, expression of each of the AT1R amber mutants transiently transfected into HEK293T cells in the absence (−) or presence (+) of 0.5 mm azF, as determined through antibody detection of the C-terminal RlucII epitope. Data are representative blots of three independent experiments (upper panel, including both sets of Western blots). Quantifications from blots are represented as mean ± S.E. (error bars) of the optical density of the band of three independent experiments and expressed as percentage of WT-AT1R (dashed line; lower panel). N.D., not detected. One-way ANOVA followed by Dunnett's multiple comparison tests was performed: *, p < 0.05; **, p < 0.01.