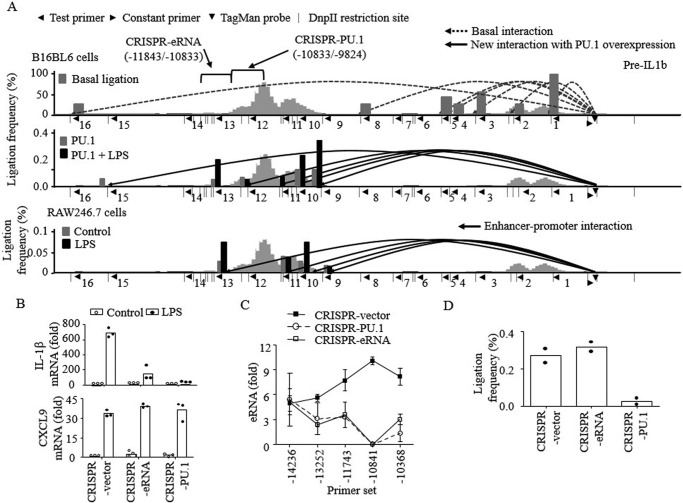
Figure 5.
PU.1 induces E2-promoter looping formation prerequisite for E2 eRNA and IL-1β mRNA transcription. A, nuclei were extracted from WT B16 cells (top), B16 cells transfected with PU.1 with or without LPS stimulation (6 h; middle), and RAW macrophages with or without LPS stimulation (3 h; bottom). Genomic DNAs were purified after digestion with DpnII and subsequently ligated and de-cross-linked. Proximal localization of DNA fragments of E2 and IL-1β promoter was analyzed by chromatin conformation capture assay, followed by TaqMan probe qPCR as described under “Experimental procedures.” The numbers below the lines (1–16) indicate the test (reverse orientation) primers used to probe the digested DNA fragments. Each test primer (◀) was utilized in combination with the promoter-recognizing constant (forward orientation) primer (▶) and TaqMan probe (▾). The dotted lines (top) indicate basal ligation between the promoter and DNA fragments detected in all samples regardless of PU.1 expression or LPS stimulation. The solid lines represent newly detected interactions after PU.1 expression in B16 cells (middle) or RAW cells with/without LPS stimulation (bottom). The bars (gray, untreated; black, LPS-treated) in each diagram represent the frequencies of ligation, compared with the ligation frequency of the promoter window and adjacent DNA fragment (primer 1). B–D, E2-deleted (CRISPR-eRNA; Δ11843–10833) and PU.1-binding region–deleted (CRISPR-PU.1; Δ10833–9824) B16 cells were generated using the CRISPR/Cas9 gene-editing system as described under “Experimental procedures” and in Fig. S2. CRISPR vector, CRISPR-eRNA, and CRISPR-PU.1 cells were transfected with pcDNA3-HA-PU.1 plasmid for 48 h as described under “Experimental procedures.” Cells were then stimulated with LPS (100 ng/ml) for 6 h, and transcription of IL-1β and CXCL9 mRNAs (B) and E2 eRNA (C) was examined by RT-qPCR. The proximal localization of E2 and IL-1β promoter was analyzed, and cumulative values of primer sets 9, 10, and 11 were plotted (D). Data are expressed as means ± S.D. (error bars) (B and C, n = 3; D, n = 2).