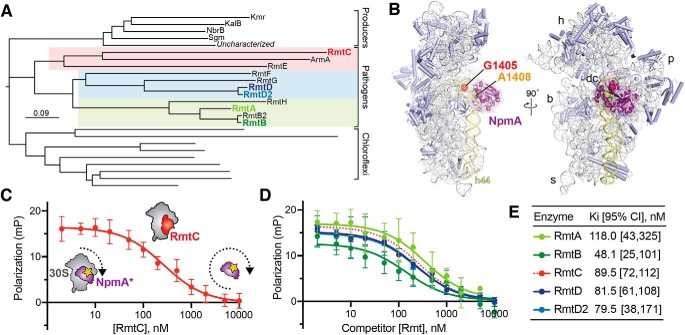
Figure 1.
The m7G1405 methyltransferase family and binding site on 30S subunit. A, phylogenetic tree of m7G1405 methyltransferase enzymes including acquired genes in gammaproteobacterial and Gram-negative pathogenic bacteria, aminoglycoside producing bacteria, and uncharacterized homologs belonging to the chloroflexi. Pathogen-associated genes (color-coded regions) are further divided into three subclades represented in this work by RmtA/RmtB, RmtD/RmtD2, and RmtC. B, structure of the bacterial 30S subunit bound to NpmA (purple) (PDB code 4OX9) showing the proximity of nucleotides G1405 (red) and A1408 (orange) at the top of h44 (yellow) in the ribosome decoding center (dc). Other 30S features indicated are the head (h), body (b), platform (p), and spur (s). C, schematic of the competition FP assay for 30S-methyltransferase binding using the NpmA* probe (purple) and application to RmtC binding (red). At low competitor concentration (left of plot), high FP signal arises because of NpmA* interaction with 30S; displacement of the probe by RmtC results in lower FP signal from the free probe (right of plot). D and E, competition FP binding experiments with NpmA* and five different unlabeled WT Rmt enzymes. The RmtC curve in D (red dotted line) is the same as in C and is shown for comparison. Binding affinities (Ki) and associated 95% confidence interval were obtained from fits to the data shown in C and D. Error bars represent S.D. of the measurements.