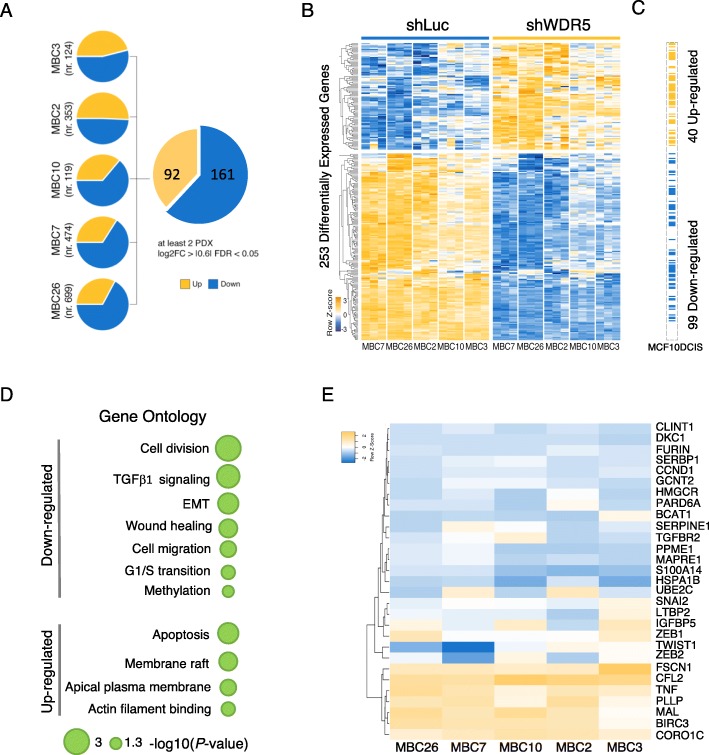
Fig. 2.
WDR5 regulates gene expression in breast cancer. RNA-seq was performed in 5 PDXs upon WDR5 silencing. a Pie charts showed differentially expressed genes (DEGs) obtained by pairwise analysis in each PDX comparing control (shLuc) and shWDR5 MBC PDXs cells. Genes were identified as DEGs when the following criteria were met: log2 fold change (FC) ≥ |0.6|, false discovery rate (FDR) < 0.05, and expression > 0.5 RPKM. DEGs in common at least in two PDXs were considered for further analysis. b Heatmap showed normalized expression values after removing batch effects of 253 DEGs identified as mentioned above (row scaled, z-score). c Corresponding genes which expression was significantly modified in MCF10DCIS cell line upon WDR5 silencing are reported. Blue and orange colors indicate down- and upregulated genes, respectively. d Representative Gene Ontology (GO) terms enriched in DEGs of PDXs are shown. For complete list refer to Supplementary Table S3. e Hierarchical clustering (Euclidean distance, complete linkage) of log2FC of expression levels (expressed as z-score values) of genes involved in enriched functions due to WDR5 silencing in five different PDXs. Listed genes are included in cell cycle progression (Cell division, G1/S transition), epithelial-to-mesenchymal transition (EMT), TGFβ1 signaling, wound healing, cell migration, and cell shape (i.e., actin binding and membrane raft) (see text). Log2FC of each gene was calculated as ratio of gene expression of shWDR5 to shLuc (control) PDX samples (done in triplicate)