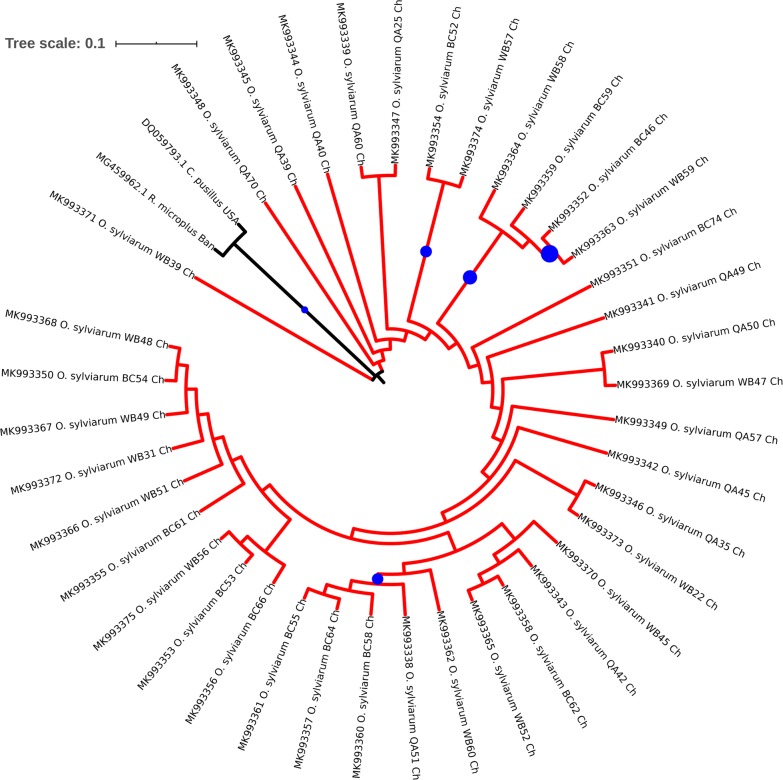
Fig. 6.
The evolutionary history was inferred using the Neighbor-Joining method based on Chinese isolates generated in this study. Sequence data generated in this study are presented in QA, BC, WB with GenBank accession numbers, species and country names. Bootstrap values (> 70%) are displayed as circles on the branches. Circle sizes at nodes are the proportion of 500 bootstrap resamplings that support the topology shown. The optimal tree with the sum of branch length of 0.43874121 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. This analysis involved 40 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). Evolutionary analyses were conducted in MEGA. Clades are coloured according to the species: Ornithonyssus sylviarum (mite: red), Chlaenius pusillus (insect: black) and Rhipicephalus microplus (tick: black). Abbreviations: Ch, China; Ban, Bangladesh; USA, United States of America