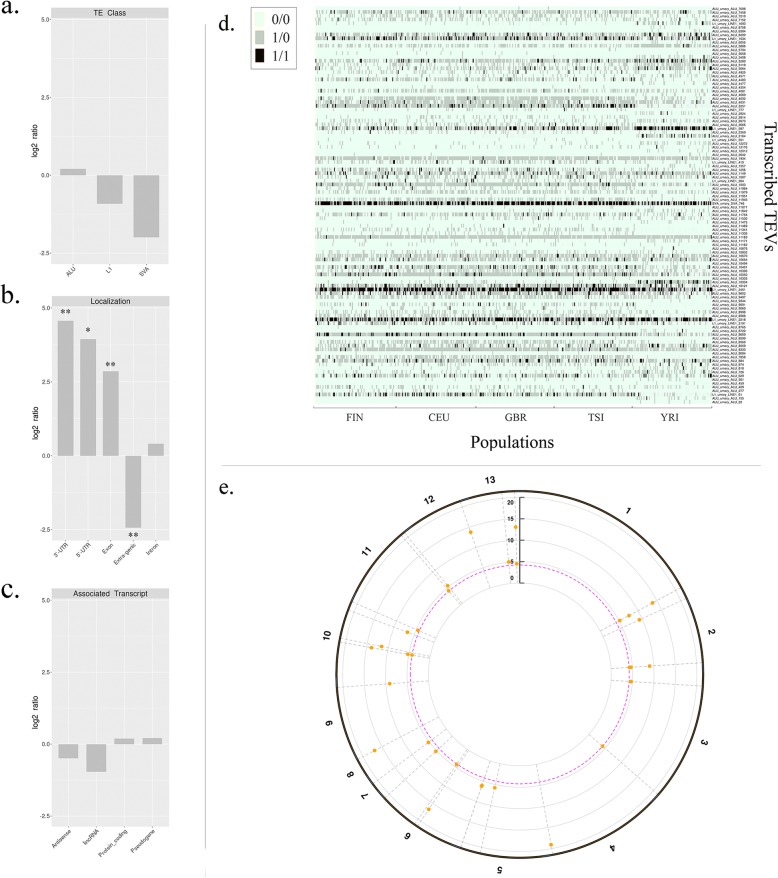
Fig. 5.
Putatively transcribed TEVs: a. Log2 of the ratio between the percentage of RNA-TEV and total TEV for each TEV class; b. Log2 of the ratio between the percentage of RNA-TEV and total TEV embedded in different genomic locations (Fisher test, ** = p-value < 0.0001, * = p-value < 0.001); c. Log2 of the ratio between the percentage of RNA-TEV associated to different gene types and the percentage composition of the total transcriptome for each transcript type; d. heatmap representing the distribution of RNA-TEVs among the individuals of the 5 analysed populations, the population are displayed on the x-axis, while the RNA-TEV are plotted on the y-axis e. Circular Manhattan plot representing the RNA-TEV associated to an altered gene expression, genomic coordinates are plotted on the x-axis while the negative log10 of the p-value associated to each RNA-TEV-gene association is displayed on the y-axes, significant associations are represented as orange dots above the significance line (FDR = 0.05)