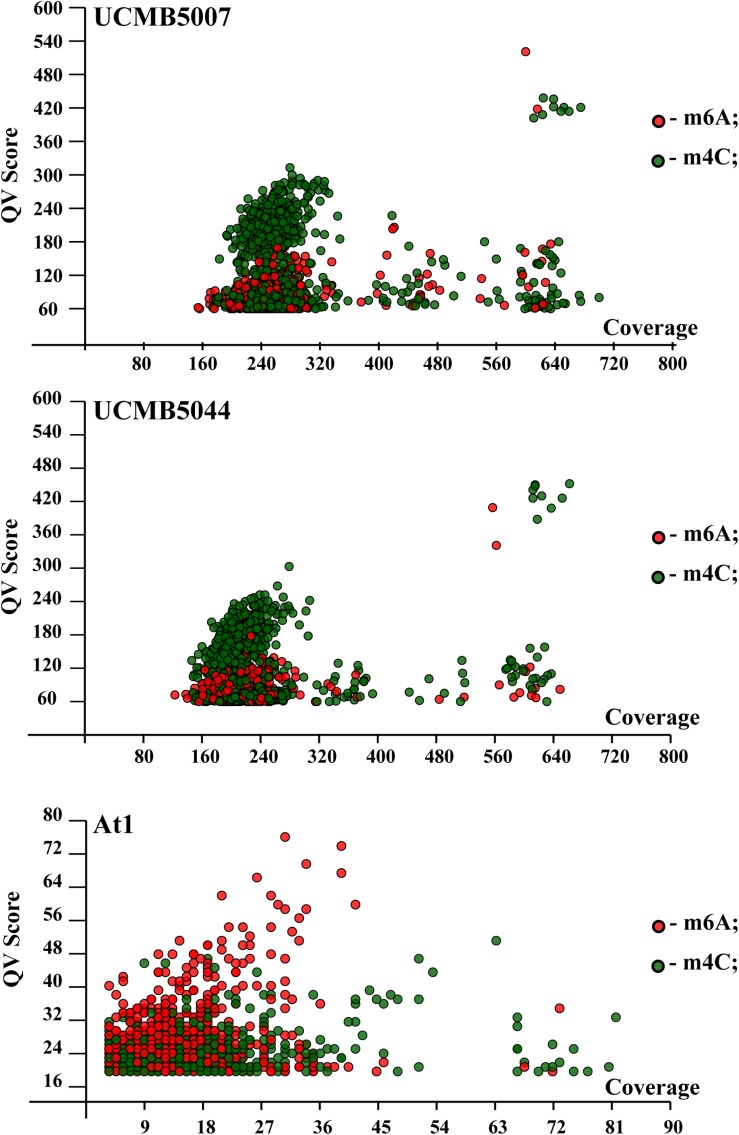
FIGURE 7.
QV modification score vs. coverage scatterplots calculated for UCMB5007, UCMB5044 and At1 genomes. Base modification dots are plotted by the corresponding QV scores and coverage values estimated by SMRT Link ipdSummary tool based on the alignments of PacBio reads against the reference sequences. Methylation types are denoted by dots of different styles, as shown in the legend. The m4C sites may correspond to both types of cytosine methylation at 4th and 5th carbon atoms.