Abstract
Purpose
Cancer ascertainment using large-scale electronic health records is a challenge. Our aim was to propose and apply a structured approach for evaluating multiple candidate approaches for cancer ascertainment using colorectal cancer (CRC) ascertainment within the US Department of Veterans Affairs (VA) as a use case.
Methods
The proposed approach for evaluating cancer ascertainment strategies includes assessment of individual strategy performance, comparison of agreement across strategies, and review of discordant diagnoses. We applied this approach to compare three strategies for CRC ascertainment within the VA: administrative claims data consisting of International Classification of Diseases, Ninth Revision (ICD9) diagnosis codes; the VA Central Cancer Registry (VACCR); and the newly accessible Oncology Domain, consisting of cases abstracted by local cancer registrars. The study sample consisted of 1,839,043 veterans with index colonoscopy performed from 1999 to 2014. Strategy-specific performance was estimated based on manual record review of 100 candidate CRC cases and 100 colonoscopy controls. Strategies were further compared using Cohen’s κ and focused review of discordant CRC diagnoses.
Results
A total of 92,197 individuals met at least one CRC definition. All three strategies had high sensitivity and specificity for incident CRC. However, the ICD9-based strategy demonstrated poor positive predictive value (58%). VACCR and Oncology Domain had almost perfect agreement with each other (κ, 0.87) but only moderate agreement with ICD9-based diagnoses (κ, 0.51 and 0.57, respectively). Among discordant cases reviewed, 15% of ICD9-positive but VACCR- or Oncology Domain–negative cases had incident CRC.
Conclusion
Evaluating novel strategies for identifying cancer requires a structured approach, including validation against manual record review, agreement among candidate strategies, and focused review of discordant findings. Without careful assessment of ascertainment methods, analyses may be subject to bias and limited in clinical impact.
INTRODUCTION
Increasing availability of large-scale electronic health records (EHRs) has great promise for enabling groundbreaking epidemiologic and quality improvement work and is particularly important for cancer research. Indeed, a recent report issued by the President’s Cancer Panel called for development of health information technologies (including through leveraging EHRs) to use learning health care systems to support continuous improvement in care across the cancer continuum and to use health information technologies to enhance cancer surveillance.1
The first steps in any initiative to leverage EHRs for epidemiologic research and quality improvement must include identifying a robust approach to cancer ascertainment. However, ascertainment of incident cancer derived from usual health care resources is a major challenge. Administrative claims data have been widely used for cancer ascertainment, but these may be subject to misclassification.2-4 For example, an International Classification of Diseases, Ninth Revision (ICD9) diagnosis code for colorectal cancer (CRC) may be associated with a colonoscopy performed for a patient based on clinical suspicion of cancer, even though CRC was excluded during the same evaluation episode.5 Sensitivity of administrative claims data for incident CRC has been reported to be as low as 72%.6,7 Cancer registries may also be considered, but data are not usually linked to usual health care data and are under-reported from non–hospital-based settings.1 Novel approaches that leverage information collected as part of usual care, such as pathology data, cases abstracted by local registrars, or cases identified through application of natural language processing algorithms, offer exciting potential but require careful validation and assessment.8 Realizing the full promise of novel strategies requires such methodologic work because large sample size cannot immunize against potential bias. A structured approach must be taken to validate approaches used to ascertain cancer, because failing to do so may result in biased epidemiologic research and incomplete quality improvement efforts.
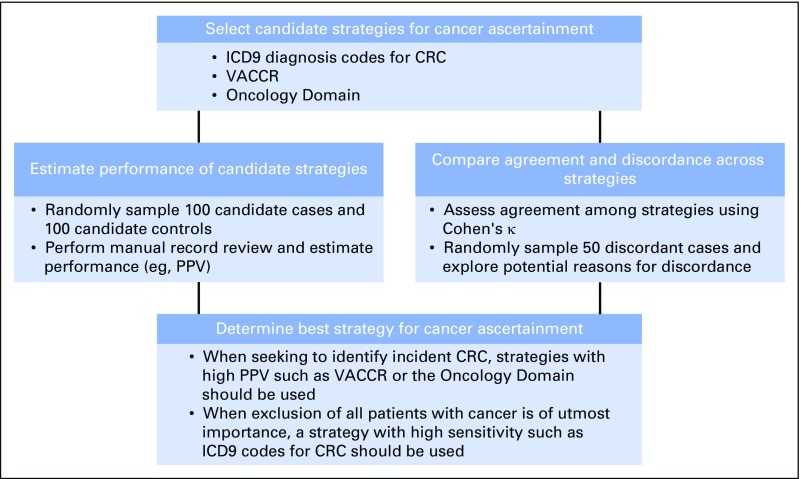
Herein we propose a structured approach for evaluating cancer ascertainment strategies derived from large-scale EHR data, including: assessment of individual strategy performance (eg, positive predictive value [PPV]), comparison of agreement across strategies, and review of discordant diagnoses (Fig 1). In this report, we apply this approach to compare three candidate strategies for CRC ascertainment within the US Department of Veterans Affairs (VA) as a use case. The approach may serve as a model for evaluating cancer ascertainment strategies derived from EHRs for epidemiologic research and quality improvement initiatives.
Fig 1.
Structured approach for evaluating strategies for cancer ascertainment. A structured approach to evaluate cancer ascertainment strategies, as well as specific application of the approach to the use case of colorectal cancer (CRC) ascertainment within the Department of Veterans Affairs are presented. ICD9, International Classification of Diseases, Ninth Revision; PPV, positive predictive value; VACCR, Veterans Affairs Central Cancer Registry.
METHODS
Overview
We conducted a retrospective comparison of CRC diagnoses ascertained by the three data sources. The study sample consisted of veterans who had undergone colonoscopy in the VA. Performance of each strategy was determined by manual record review of a subsample of 100 candidate CRC cases and 100 candidate controls for which colonoscopy was performed in the VA and summarized. Agreement among strategies and careful review of discordant findings were conducted to determine etiology of discordance. The primary outcome was CRC diagnosis within 30 days of index colonoscopy.
Study Setting and Data Sources
The VA is the largest integrated health care system in the United States. A wide array of usual care data have been collected since 1999 and reflect care of almost 6 million veterans annually.9 Available data include EHRs as well as complimentary sources such as cancer registries.10 As such, the VA offers one of the largest resources for cancer research in the United States. Data are housed within the VA Informatics and Computing Infrastructure, which allows secure access to national VA data. The Corporate Data Warehouse (CDW) is a large data repository and contains both clinical data (medications, laboratory tests, and pathology results) and administrative claims (diagnoses and procedure codes).11 Within the CDW, ICD9 diagnosis codes recorded during any inpatient or outpatient setting represent one potential source for cancer ascertainment.
The VA Central Cancer Registry (VACCR) has served as the gold standard of cancer ascertainment for the last decade.12 However, because of constrained resources, a significant time lag exists between case abstraction at local VA sites and final inclusion within the registry. Furthermore, data requests place a significant burden on already limited registry resources and are associated with significant time from request to data provision.
The Oncology Domain has recently become available to researchers within the CDW (Appendix Fig A1). Oncology Domain files represent data abstracted at the local level by cancer registrars. These data are used by VACCR for creation of finalized registry data. Data available within the Oncology Domain have not been previously cleaned or aggregated, but we postulate that restricting analyses of Oncology Domain data to those marked as having complete abstract status at the local level may result in data that mimic the quality of VACCR data, particularly for ascertainment of incident cancer, because local registrars have abstracted cases. Because these data are more easily accessible to VA researchers and quality improvement leaders, are immediately available after local registrar abstraction, and contain detail similar to VACCR data, the Oncology Domain may be a promising, practical resource for cancer ascertainment.
Study Sample and Candidate Case Ascertainment
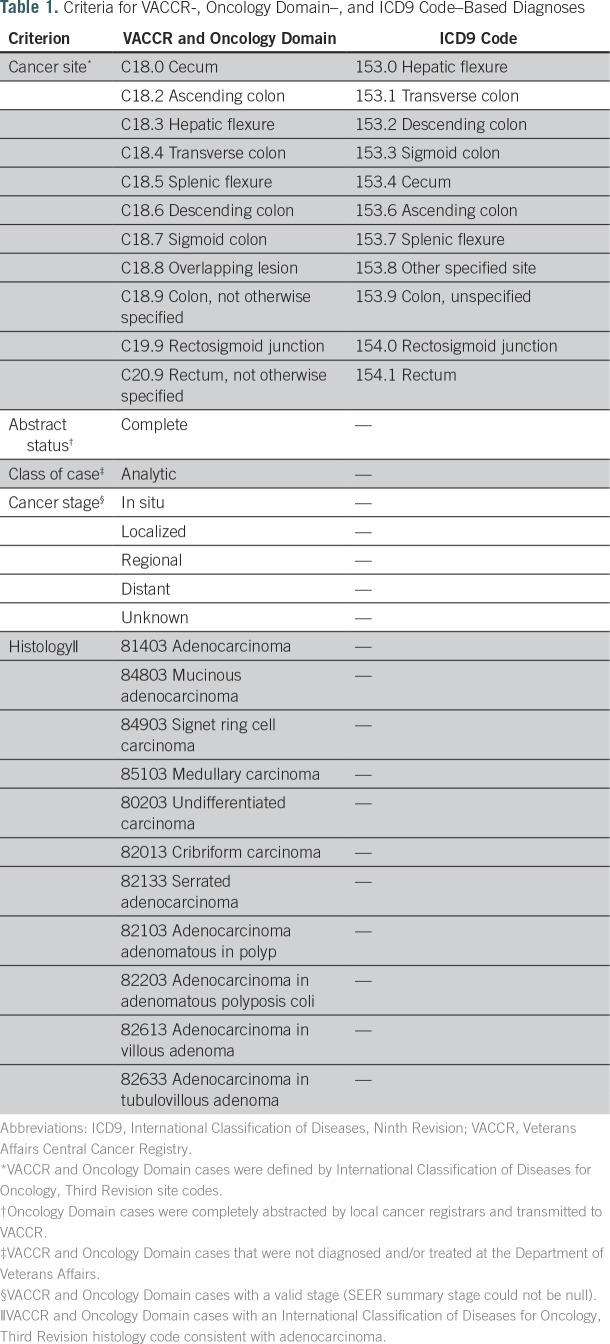
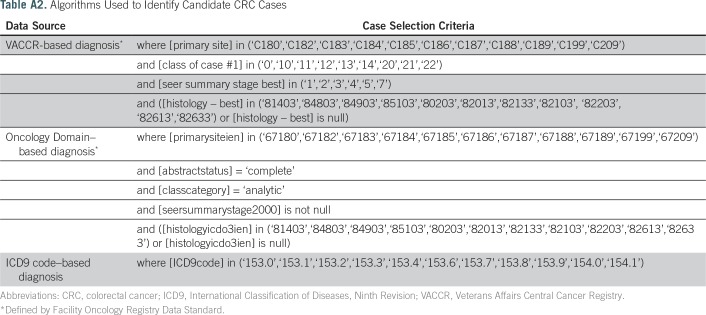
The study sample consisted of veterans with at least one Current Procedural Terminology code for colonoscopy performed in an inpatient or outpatient setting from January 1, 1999, to December 31, 2014 (Appendix Table A1 lists codes used). To identify candidate cases, we created three algorithms for CRC diagnosis based on VACCR, the Oncology Domain, and ICD9 diagnosis codes (Appendix Table A2 lists algorithms used). Cases from VACCR and the Oncology Domain were defined by International Classification of Diseases for Oncology, Third Revision site codes. For each CRC definition, we selected the first instance recorded. Prevalent CRC was defined as occurring up to 6 months before or after the index colonoscopy. Additional exclusion criteria, including abstract status, class of case, cancer stage, and histology, are summarized in Table 1.
Table 1.
Criteria for VACCR-, Oncology Domain–, and ICD9 Code–Based Diagnoses
Statistical Analysis
For individuals who fit under each definition of CRC, patient characteristics and features of cancer presentation were abstracted and summarized. Demographics (sex, race/ethnicity) were ascertained from the CDW. Age at diagnosis was calculated as the difference between date of birth and date of presumed cancer diagnosis. Features of cancer presentation (primary site, cancer stage) were obtained from VACCR and the Oncology Domain. Primary site was split into proximal (C18.0, C18.2 to C18.5), distal (C18.6 to C18.7), rectal (C19.9, C20.9), and other (C18.8 to C18.9). Cancer stage was characterized as in situ, localized, regional, distant, or unknown.
Validation of candidate strategies.
We developed a structured approach to independently validate each cancer ascertainment strategy in which we created an algorithm for each potential resource, applied the algorithm to our study sample, and estimated CRC prevalence based on each ascertainment strategy. For each CRC algorithm, we randomly sampled 100 candidate CRC cases and 100 candidate controls from our study sample of individuals who had undergone colonoscopy but did not meet criteria for CRC diagnosis. As such, our sampling resulted in three independent sample sets: 100 VACCR-based cases and 100 colonoscopy controls, 100 Oncology Domain–based cases and 100 controls, and 100 ICD9-based cases and 100 controls. For each sample set, reviewers (A.E. and R.B.) were blinded to whether each patient was a case or control and searched records for evidence of CRC diagnosis within the EHR.13
Performance of each algorithm was estimated by PPV and negative predictive value (NPV) using manual record review as the gold standard. Sample size was based on the 100(1 − α/2)% one-sided confidence lower bounds for PPV and NPV. Bonferroni correction was used for multiple comparison adjustment to ensure an overall confidence of 95%. We postulated that if estimated PPV and NPV reached ≥ 95%, the 97.5% one-sided confidence lower bounds for PPV and NPV would be > 0.90, and we could confidently conclude that the true PPV and NPV were > 90% (manuscript in preparation). PPV and NPV for each algorithm were then combined with estimated prevalence to calculate sensitivity and specificity.
Agreement and discordance across candidate strategies.
To assess whether multiple strategies could improve accuracy, we estimated agreement across strategies and randomly sampled discordant findings to explore potential reasons for discordance. Agreement was evaluated by Cohen’s κ14,15 and defined as (Po − Pe)/(1 − Pe), where Po is the observed proportion of individuals for which two methods agree and Pe is the probability that two methods agree by chance, based on the observed case and control classifications of each method. κ is > 0 if the observed agreement exceeds the proportion expected by chance and reaches its maximum value of 1 when two methods reach perfect agreement. CIs were calculated for κ, and Bonferroni correction was applied for three agreement measures with an overall confidence of 95%. κ > 0.80 represents almost perfect agreement, 0.61 to 0.80 represents substantial agreement, and 0.41 to 0.60 represents moderate agreement.14
There were six types of discordance: VACCR positive versus Oncology Domain negative, Oncology Domain positive versus VACCR negative, VACCR positive versus ICD9 negative, Oncology Domain positive versus ICD9 negative, ICD9 positive versus VACCR negative, and ICD9 positive versus Oncology Domain negative. We randomly sampled 50 cases for each type of discordance and conducted focused record reviews to determine presence or absence of CRC and potential reason for discrepancy.
RESULTS
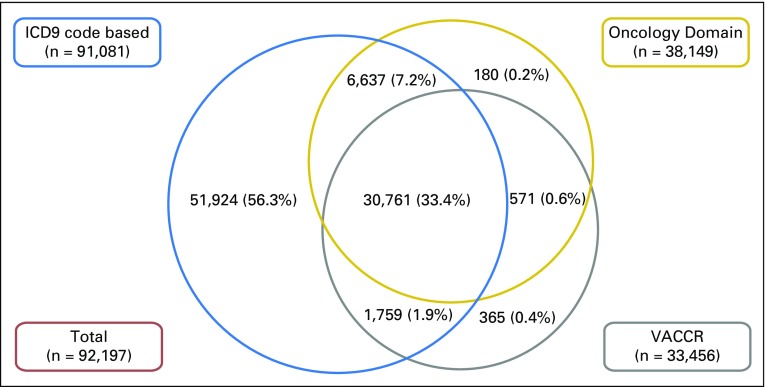
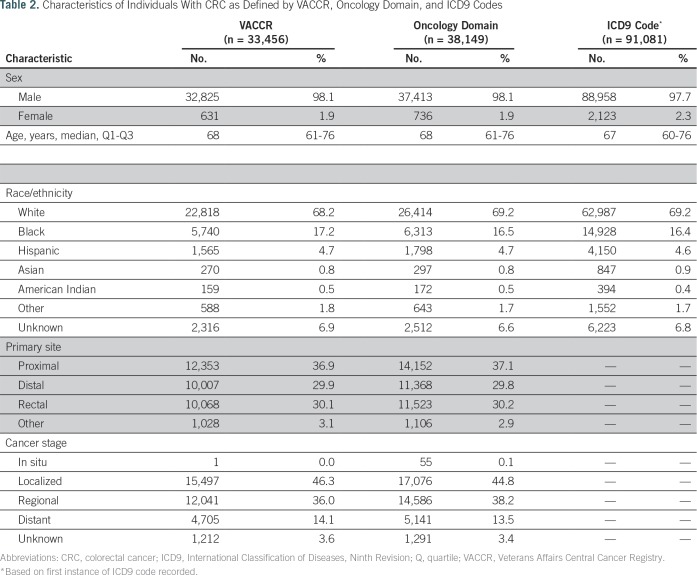
From a study sample of 1,839,043 veterans with index colonoscopy from 1999 to 2014, 92,197 met criteria for CRC diagnosis based on one or more of our candidate strategies. Figure 2 depicts the overlap in candidate CRC diagnoses across strategies. VACCR and the Oncology Domain had high overlap, such that a small proportion were identified as candidate cases by one but not the other source. Although ICD9-based ascertainment of candidate cases captured nearly all cases in VACCR or the Oncology Domain, a high proportion (56.3%) of ICD9-based candidate cases were not found in VACCR or the Oncology Domain. Table 2 lists summary statistics for individuals with suspected CRC. Most patients were white men with a median age of 68 years. On the basis of VACCR and the Oncology Domain, CRC cases most commonly originated in the proximal colon (VACCR, 36.9%; Oncology Domain, 37.1%) and were localized (VACCR, 46.3%; Oncology Domain, 44.8%).
Fig 2.
Overlap in candidate colorectal cancer (CRC) diagnoses across ascertainment strategies. A total of 92,197 individuals met at least one CRC definition. Veterans Affairs Central Cancer Registry (VACCR) and the Oncology Domain had high overlap, such that a small proportion of all individuals were identified as candidate CRC cases by one but not the other source. Although International Classification of Diseases, Ninth Revision (ICD9) –based ascertainment of candidate cases captured nearly all cases in VACCR or the Oncology Domain, 56.3% of ICD9-based candidate cases were not found in VACCR or the Oncology Domain. NOTE. Proportions are not to scale for ease of presentation.
Table 2.
Characteristics of Individuals With CRC as Defined by VACCR, Oncology Domain, and ICD9 Codes
Validation of CRC Ascertainment Strategies
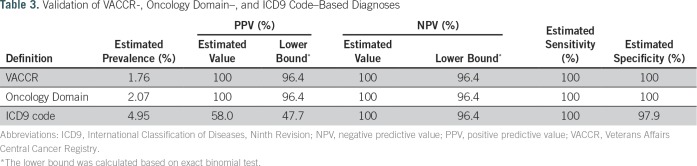
Both VACCR- and Oncology Domain–based methods were estimated to have near perfect PPV, NPV, sensitivity, and specificity when compared against manual record review as the gold standard (Table 3). The ICD9 code–based strategy was less robust. Although sensitivity, specificity, and NPV were high, PPV was suboptimal at 58%. Among the ICD9-based cases that did not have evidence of CRC upon manual record review (n = 42), 27 were issued the code for suspicion of CRC that was later ruled out by colonoscopy, and 15 reflected prior history of CRC rather than diagnosis around the time of the colonoscopy procedure.
Table 3.
Validation of VACCR-, Oncology Domain–, and ICD9 Code–Based Diagnoses
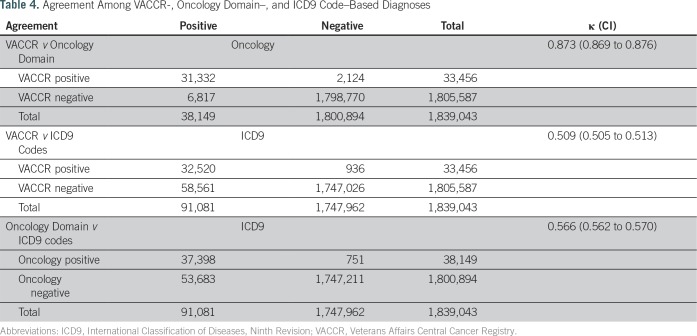
Agreement Among Ascertainment Strategies and Evaluation of Discordant Findings
Table 4 summarizes the level of agreement among our three CRC ascertainment strategies, using all data available (N = 1,839,043 with colonoscopy). Although VACCR- and Oncology Domain–based diagnoses demonstrated almost perfect agreement (κ, 0.87), the ICD9-based strategy had only moderate agreement with VACCR (κ, 0.51) and the Oncology Domain (κ, 0.57). The main reason for this discordance was that there were many more ICD9 code–based diagnoses than VACCR- or Oncology Domain–based diagnoses (Fig 2). For example, although 32,520 cases (35.7%) were consistent with CRC based on both VACCR and ICD9 criteria, the remaining 58,561 cases (64.3%) did not have VACCR data consistent with the ICD9-based approach.
Table 4.
Agreement Among VACCR-, Oncology Domain–, and ICD9 Code–Based Diagnoses
To assess accuracy of each strategy for identifying CRC, we reviewed a random sample of discordant cases across strategies to determine presence or absence of CRC and etiology of discordance. All cases that were in VACCR but not in the Oncology Domain (or vice versa) had evidence of a CRC diagnosis at time of index colonoscopy. Similarly, all cases that were in VACCR or the Oncology Domain but did not have an ICD9 code–based diagnosis were confirmed to have incident CRC as well. However, only 15% of cases that had an ICD9 code but not a VACCR- or Oncology Domain–based diagnosis had evidence of CRC at index colonoscopy. The remaining cases were issued either the code for suspicion of CRC that was later ruled out by colonoscopy (38%) or the code reflecting prior history of CRC rather than diagnosis around the time of the colonoscopy procedure (47%).
DISCUSSION
Cancer ascertainment using large-scale EHRs is a challenge. Methods for ascertainment should be chosen based on accuracy, accessibility, research questions under study, and purpose of ascertaining diagnoses. In this work, we proposed a structured approach for evaluating cancer ascertainment strategies using large-scale EHR data and subsequently implemented our approach to compare three candidate strategies for ascertainment of CRC within the VA as a use case.
All three strategies showed high sensitivity and specificity for incident CRC. However, the ICD9 code–based approach had a much lower PPV than the approaches based on VACCR or the newly accessible Oncology Domain. Specifically, PPV for ICD9 code–based strategy was just 58% in comparison with 100% for VACCR and the Oncology Domain. In contrast, our evaluation of the agreement of VACCR-, Oncology Domain–, and ICD9 code–based diagnoses suggests that VACCR and the Oncology Domain do have limitations. Specifically, 15% of ICD9-positive but VACCR- or Oncology Domain–negative cases were confirmed to have CRC at index colonoscopy.
Application of multiple approaches for evaluating cancer ascertainment strategies allows us to carefully assess the strengths and weaknesses of candidate approaches (Fig 1). For example, our data suggest that for research questions seeking to identify incident CRC with high PPV, VACCR or the Oncology Domain should be used preferentially over ICD9-based criteria. In contrast, in situations where exclusion of all patients with baseline presence or history of CRC is of paramount importance, ICD9 codes should be considered as an adjunct to VACCR or the Oncology Domain. Our results also provide a cautionary example of why multiple approaches must be taken toward validating the accuracy of rare predictors or outcomes of interest. If we had only relied on a random sample of cases and controls, we would have assumed near-perfect sensitivity for VACCR and the Oncology Domain. Indeed, our finding that 15% of ICD9-positive but VACCR- or Oncology Domain–negative cases had a cancer diagnosis suggest that the sensitivity of these sources for cancer ascertainment can still be improved.
Our findings support the use of a structured, hybrid approach to evaluating candidate strategies implemented for EHR data (Fig 1). Specifically, by considering multiple strategies and comparing outputs using a structured approach including validation and agreement as well as record review of discordant cases, the strengths and limitations of each strategy can be well understood. We postulate that using large data sets from EHRs without such work might risk incorrect or underascertainment that could go unrecognized.
Several limitations should be considered when interpreting our work. First, we used a relatively simple ICD9 code–based strategy (first instance recorded). Others have considered other approaches (eg, multiple codes over time) to improve specificity.2,3 Development and validation of a more complex strategy were beyond the scope of this work. Second, we focused on validating approaches for identifying CRC found at the time of index colonoscopy. Indeed, some of the ICD9-based positive diagnoses were in individuals who had a history of CRC. This speaks to the importance of validating the diagnostic approach for the purpose of the research under way; in our case, we were mainly interested in identification of individuals with incident cancer at time of index colonoscopy. Third, caution should be taken in generalizing our findings to other cancer diagnoses. Future work should test whether our approach can be applied in other cancer types, in addition to using other data resources. Fourth, VACCR and Oncology Domain data are nonindependent, because in our system, Oncology Domain data inform final VACCR data. Although we validated each ascertainment strategy separately with independent random samples, potential for data correlation does exist. Strengths of this work include use of multiple approaches to assess strengths and weakness of each strategy and use of data from the largest integrated health system in the United States.
For researchers and quality improvement leaders interested in cancer research within the VA, our findings suggest that the Oncology Domain may be considered as an alternative source for cancer ascertainment. Indeed, we found 6,637 additional CRC cases in the Oncology Domain that were not in VACCR. This is the first report to our knowledge that has validated this newly accessible resource. Our results suggest that the Oncology Domain may continue to provide a valid resource for ascertaining cancer diagnoses.
Beyond the VA, our work suggests that a structured approach must be taken to evaluate strategies for identifying cancer outcomes and recommends considering validation using random sample record review, as well as evaluating agreement among candidate strategies. Additionally, we postulate that strategic sampling and manual record review of cases where definitions offer discordant conclusions in particular may be helpful in understanding the strengths and limitations of novel approaches, particularly those designed to identify rare predictors and outcomes.
ACKNOWLEDGMENT
We thank the Office of Patient Care Services for maintaining and distributing the Veterans Affairs (VA) Central Cancer Registry (VACCR), and the VA Informatics and Computing Infrastructure for maintaining and distributing the Oncology Domain within the Corporate Data Warehouse. We also acknowledge Olga V. Patterson for her assistance in creating our study sample, Daniel Denhalter for helping set up the ChartReview tool, and Pat Coke for patiently guiding us through the entire VA cancer data abstraction process.
Appendix
Table A1.
CPT Procedure Codes Used to Create Study Sample
Table A2.
Algorithms Used to Identify Candidate CRC Cases
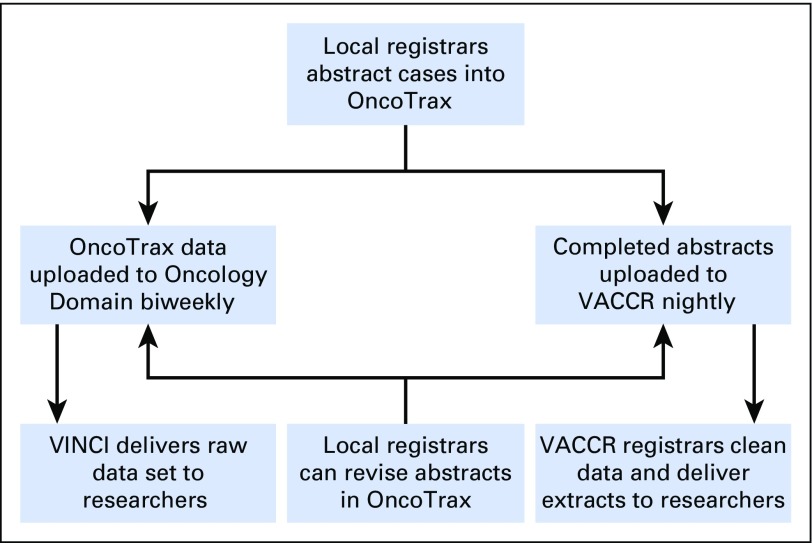
Fig A1.
Overview of the Veterans Affairs (VA) cancer data abstraction process. Local registrars from VA medical centers across the country manually abstract cases into OncoTrax. Completed abstracts are transmitted to the VA Central Cancer Registry (VACCR) nightly. VACCR registrars clean and aggregate the data and deliver research-grade extracts to researchers upon request. OncoTrax data have recently become available through tables known as the Oncology Domain. OncoTrax data are uploaded to the Oncology Domain biweekly. The VA Informatics and Computing Infrastructure (VINCI) delivers static data sets to researchers upon request.
Footnotes
Supported by Merit Review 5 I01 HX001574-03 (S.G., Principal Investigator [PI]) and Quality Enhancement Research Initiative 5 IP1 HX002002-03 (T.K., Project 1 PI) from the US Department of Veterans Affairs (VA) Health Services Research and Development Service of the VA Office of Research and Development.
The views expressed in this article are those of the authors and do not necessarily represent the views of the US Department of Veterans Affairs.
AUTHOR CONTRIBUTIONS
Conception and design: Ashley Earles, Lin Liu, Pat Coke, Julie Lynch, Karen Messer, Andrew J. Gawron, Tonya Kaltenbach, Samir Gupta
Collection and assembly of data: Ashley Earles, Ranier Bustamante, Pat Coke, Samir Gupta
Data analysis and interpretation: Ashley Earles, Lin Liu, Ranier Bustamante, Julie Lynch, Karen Messer, María Elena Martínez, James D. Murphy, Christina D. Williams, Deborah A. Fisher, Dawn T. Provenzale, Andrew J. Gawron, Tonya Kaltenbach, Samir Gupta
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Ashley Earles
No relationship to disclose
Lin Liu
Leadership: BetaCyte Laboratories (I)
Ranier Bustamante
No relationship to disclose
Pat Coke
No relationship to disclose
Julie Lynch
Research Funding: Genomic Health (Inst), AstraZeneca (Inst), Myriad Genetics (Inst), Boehringer Ingelheim (Inst), Astellas Pharma (Inst), CardioDx (Inst), Janssen (Inst)
Karen Messer
No relationship to disclose
María Elena Martínez
No relationship to disclose
James D. Murphy
No relationship to disclose
Christina D. Williams
No relationship to disclose
Deborah A. Fisher
No relationship to disclose
Dawn T. Provenzale
No relationship to disclose
Andrew J. Gawron
No relationship to disclose
Tonya Kaltenbach
Consulting or Advisory Role: Olympus
Travel, Accommodations, Expenses: Olympus
Samir Gupta
Consulting or Advisory Role: Exact Sciences, Guidepoint Global Consulting, BibCon, GLG Consulting
Research Funding: Polymedco
REFERENCES
- 1. President’s Cancer Panel: Improving Cancer-Related Outcomes with Connected Health: A Report to the President of the United States From the President’s Cancer Panel. https://prescancerpanel.cancer.gov/report/connectedhealth/pdf/PresCancerPanel_ConnHealth_Nov2016.pdf.
- 2.Cooper GS, Yuan Z, Stange KC, et al. : The sensitivity of Medicare claims data for case ascertainment of six common cancers. Med Care 37:436-444, 1999 [DOI] [PubMed] [Google Scholar]
- 3.Clarke CL, Feigelson HS: Developing an algorithm to identify history of cancer using electronic medical records. EGEMS (Wash DC) 4:1209, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts RJ, Stockwell DC, Slonim AD: Discrepancies in administrative databases: Implications for practice and research. Crit Care Med 35:949-950, 2007 [DOI] [PubMed] [Google Scholar]
- 5.O’Malley KJ, Cook KF, Price MD, et al. : Measuring diagnoses: ICD code accuracy. Health Serv Res 40:1620-1639, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldsbury D, Weber M, Yap S, et al. : Identifying incident colorectal and lung cancer cases in health service utilisation databases in Australia: A validation study. BMC Med Inform Decis Mak 17:23, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldi I, Vicari P, Di Cuonzo D, et al. : A high positive predictive value algorithm using hospital administrative data identified incident cancer cases. J Clin Epidemiol 61:373-379, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Benchimol EI, Manuel DG, To T, et al. : Development and use of reporting guidelines for assessing the quality of validation studies of health administrative data. J Clin Epidemiol 64:821-829, 2011 [DOI] [PubMed] [Google Scholar]
- 9. US Department of Veterans Affairs: National Centers for Veterans Analysis and Statistics: Veteran population. http://www.va.gov/vetdata/Veteran_Population.asp.
- 10.Fihn SD, Francis J, Clancy C, et al. : Insights from advanced analytics at the Veterans Health Administration. Health Aff (Millwood) 33:1203-1211, 2014 [DOI] [PubMed] [Google Scholar]
- 11.DuVall SL, Nebeker JR: VINCI: A convergence of policy and technology for enabling big data analytics in the Department of Veterans Affairs. Presented at the 22nd Annual International Conference on Computer Science and Software Engineering, Toronto, Ontario, Canada, November 5-7, 2012 [Google Scholar]
- 12.Lamont EB, Landrum MB, Keating NL, et al. : Evaluating colorectal cancer care in the Veterans Health Administration: How good are the data? J Geriatr Oncol 2:187-193, 2011. 10.1016/j.jgo.2011.02.001 [DOI] [Google Scholar]
- 13.DuVall SL, Forbush TB, Cornia RC, et al. : Reducing the manual burden of medical record review through informatics. Presented at the 30th International Conference on Pharmacoepidemiology and Therapeutic Risk Management, Taipei, Taiwan, October 24-27, 2014 [Google Scholar]
- 14.Viera AJ, Garrett JM: Understanding interobserver agreement: The kappa statistic. Fam Med 37:360-363, 2005 [PubMed] [Google Scholar]
- 15.Landis JR, Koch GG: The measurement of observer agreement for categorical data. Biometrics 33:159-174, 1977 [PubMed] [Google Scholar]