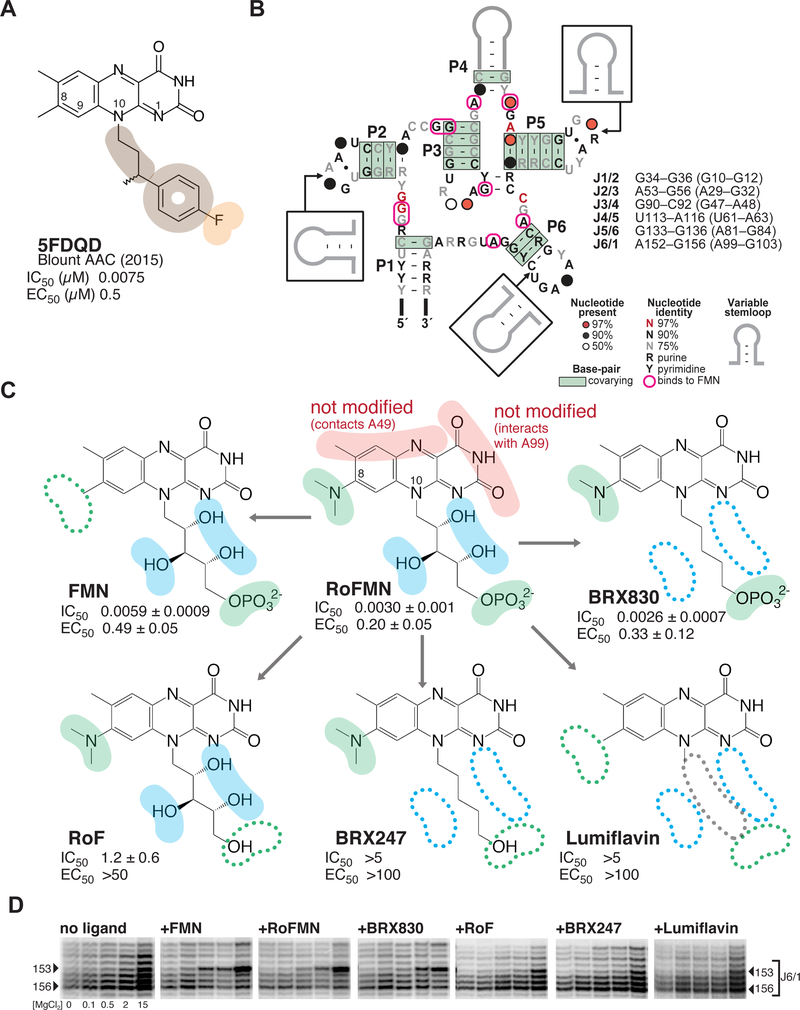
Figure 1 |. Roseoflavin mononucleotide at the center of a medicinal chemistry optimization strategy that led to the discovery of synthetic analogs with potent activity and selectivity.
(A) Chemical structure of 5FDQD (5-(3-(4-fluorophenyl)butyl)-7,8-dimethylpyrido[3,4-b]- quinoxaline-1,3(2H,5H)-dione), a potent inhibitor of the FMN riboswitch. Color-coding for functional groups introduced during SAR study: orange, negatively charged/polar group; tan, hydrophobic group. IC50, half maximal inhibitory concentration as measured by in-line probing; EC50, half maximal effective concentration in transcription termination assays.31 Note that all values for IC50 and EC50 in subsequent figures are given in units of μM. (B) Secondary structure of the FMN riboswitch showing sequence and structure conservation among bacteria. Residues that interact with flavin-bearing ligands in crystal structures are circled in pink.30, 33 The list of the six joining regions is indicated, with numbering for B. subtilis and F. nucleatum (in parenthesis). (C) Dissecting functional positions 8 and 10 of roseoflavin mononucleotide (RoFMN) over the course of a structure-activity relationship (SAR) study of the FMN riboswitch. Color-coding for functional groups: red, left unaltered during SAR study; green, primary focus of SAR study; blue, secondary focus. IC50 and EC50 calculated as for 5FDQD (see Methods; Tables S1and S2; Figure S1). (D) Comparative banding pattern of SHAPE chemical probing within the J6/1 joining region, which serves as an indicator for ligand binding.30 [MgCl2] tested were: 0, 0.1, 0.5, 2.0 and 15.0 mM. Arrowheads: residues of interest within J6/1. The gels were aligned in SAFA53–54 (full unaltered SHAPE gels shown in Figure S2).