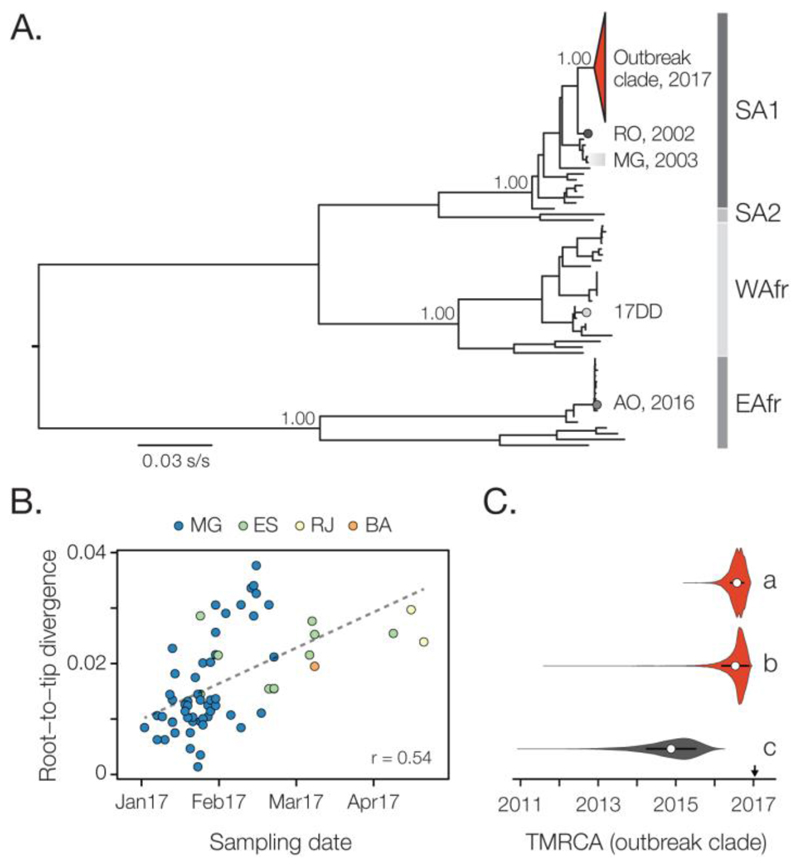
Fig. 3. Molecular phylogenetics of the Brazilian YFV epidemic.
(A) Maximum likelihood phylogeny of complete YFV genomes showing the outbreak clade (red triangle) within the SA1 genotype (see Figs. 4 and S6 for details). SA2, WAfr and EAfr indicate the South America II, West African, and East Africa genotypes, respectively. For clarity, five YFV strains introduced to Venezuela from Brazil (49) are not shown. The scale bar is in units of substitutions per site (s/s). Node labels indicate bootstrap support values. RO 2002 = strain BeH655417 from Roraima. MG 2003 = two strains from the previous YF outbreak in MG in 2003. 17DD = the vaccine strain used in Brazil. AO 2016 = YFV outbreak Angola in 2015-2016 (13). (B) Root-to-tip regression of sequence sampling date against genetic divergence from the root of the outbreak clade (see Fig. S6A). Sequences are coloured by sampling location. (C) Violin plots showing estimated posterior distributions (white circle=mean) of the time of the most common ancestor (TMRCA) of the outbreak clade. Estimates were obtained using two different datasets (grey=SA1 genotype, red=outbreak clade) and under different evolutionary models: a=uncorrelated lognormal relaxed clock (UCLN) model with a skygrid tree prior with covariates (specifically, the time series data shown in Figs. 1A-C; see Fig. S7); b=UCLN model with a skygrid tree prior without covariates; c=fixed local clock model (see Supplementary Materials).