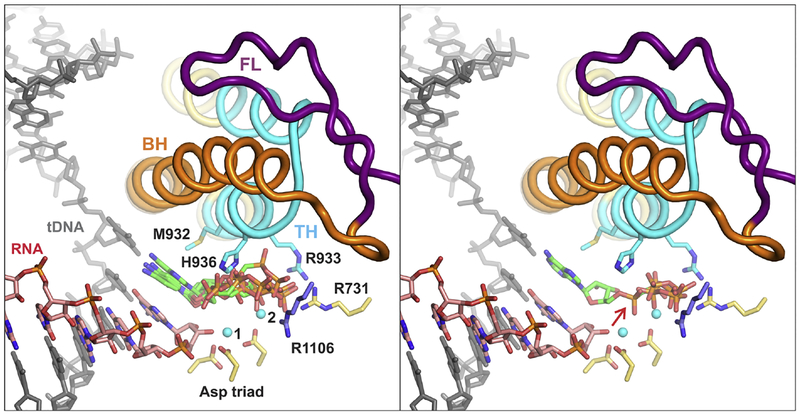
Fig. 4.
The TL folding into TH biases the substrate NTP from a pre-insertion (left panel) to the insertion pose (right panel) thereby repositioning the α-phosphate and aligning the PPi moiety inline for the attack of the 3’ OH group (right panel, red arrow). The fully closed active site depicted in both panels is drawn using the atomic coordinates of the T. thermophilus TEC with the ATP analogue AMPCPP in the insertion site (PDB ID 2O5J). In the left panel, the insertion pose of the AMPCPP was replaced with AMPCPP from T. thermophilus TEC where the active site closure is inhibited by streptolydigin (PDB ID 2PPB) and four CMPCPPs from partially closed active sites of the de novo initiation complexes (PDB IDs 4OIO, 4Q4Z, 5X22). In the right panel, the insertion pose of the AMPCPP was supplemented with two PPi molecules, one from the fully closed initially transcribing complex of E. coli RNAP (PDB ID 5IPL) and the other from the reiterative transcription complex of T. thermophilus RNAP (PDB ID 5VO8). Selected active site residues are shown as sticks; βArg1106 is colored blue, β’ residues are colored yellow or cyan. The figure was prepared using PyMOL Molecular Graphics System, Version 2.0 Schrodinger, LLC. The heterologous ligands (NTP analogues and PPi) were positioned in the closed active site using β subunits as anchors and “super” command of PyMOL.