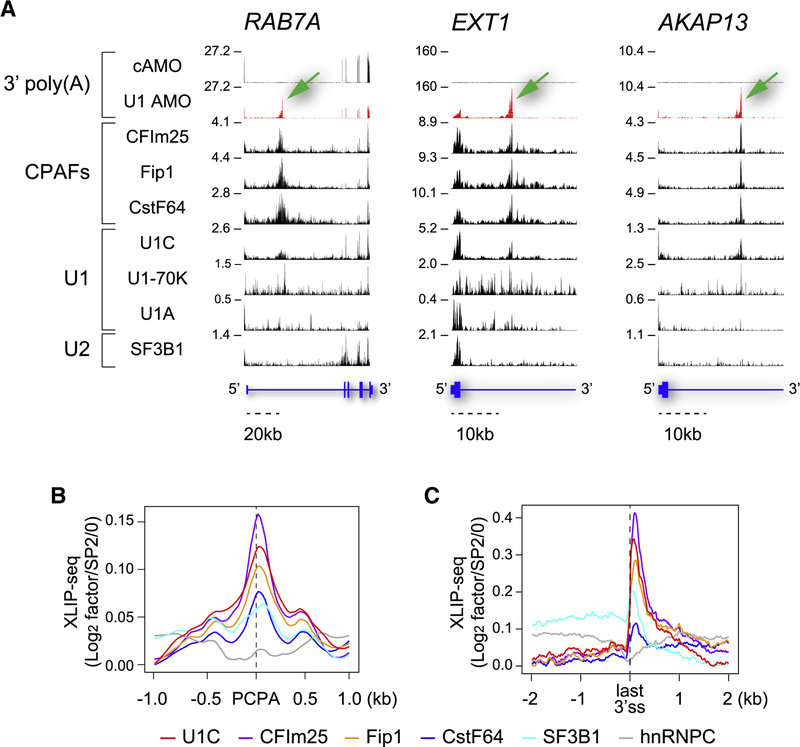
Figure 1. U1 and CPAFs co-localize at PCPA locations in introns.
(A) Genome browser views of XLIP-seq data for the indicated factors from HeLa cells transfected with U1 AMO or cAMO in select regions of representative genes (RAB7A, EXT1, and AKAP13). Non-genomic 3’-poly(A)s identified in the RNA-seq of 5min EU pulse-labeled and oligo(dT)-selected RNAs indicate the positions of PCPA elicited with U1 AMO (green arrows). In cAMO, these are located at the end of the genes, which are not included in the views shown. The Y-axis indicates reads per million (RPM) for the highest peak within the genome browser field for each sample. Annotated RefSeq gene structures are shown in blue with thin horizontal lines indicating introns and thicker blocks indicating exons (See also Figure S1). (B) Metagene plots for U1–CPAFs co-localization at PCPA sites (n=1,485). Normalized XLIP binding (log2 RPM in XLIPs over SP2/0) of the factors was rescaled using the lowest values as a baseline and were plotted around the PCPA sites within a 2kb window. (C) Metagene plots of the normalized U1–CPAFs XLIP binding for PCPA genes as shown in (B) around the last 3’ss, (n=1,469) within a 4kb window.