Figure 4.
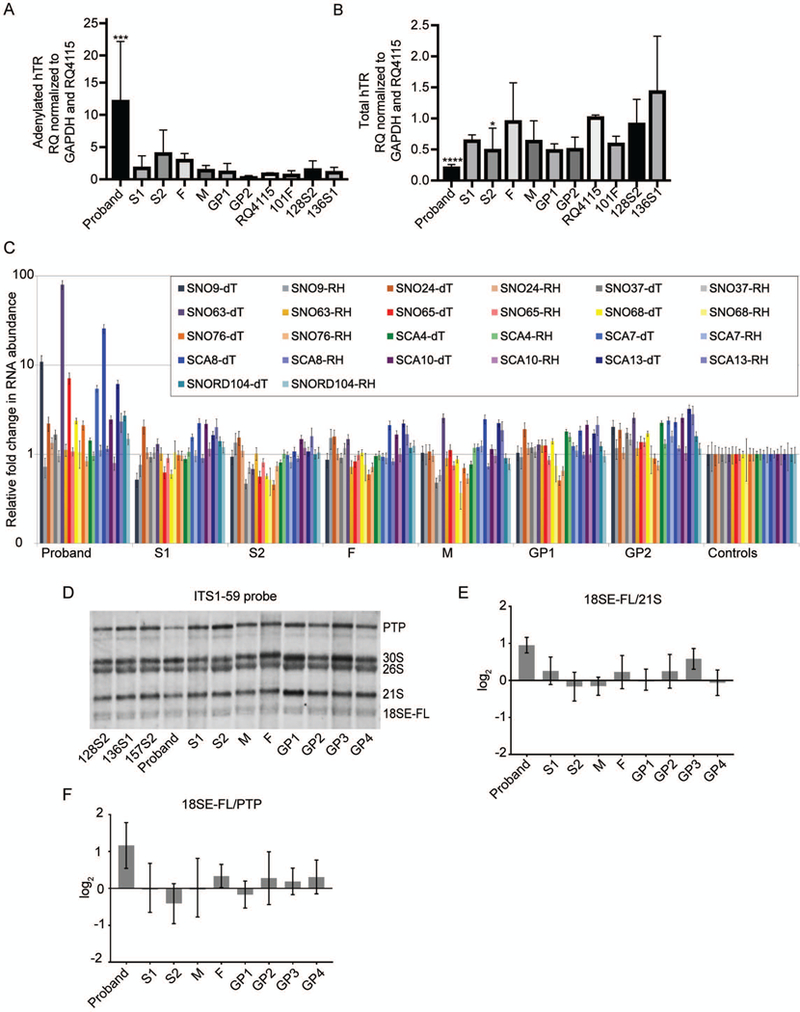
PARN RNA targets are impacted in the proband’s LCLs but not in the other BMF22 family member LCLs. (A) Analysis of adenylated hTR in BMF22 family and unrelated control (RQ4115, 101F, 128S2, and 136S1) LCLs by qRT-PCR. Complementary DNA was synthesized using oligo(dT) primer and qPCR was performed using primers to the target, hTR, or the control, GAPDH. Bars indicate mean +/− SD compared to RQ4115 with N=3–4. ***p=0.001. (B) Analysis of total hTR as in (A) except cDNA was synthesized using random hexamer primers. Bars indicate mean +/− SD compared to RQ4115 with N=3–4. *p<0.05, ****p=0.0001. (C) cDNA was synthesized from oligoadenylated RNA by using oligo(dT) primer or total RNA by using random hexamer primers. Bars indicate mean +/− SD compared to the average of the control individuals (RQ4115, 101F, and 128S2). N=3. (see text for significant p values). (D) Representative northern blot of ribosomal RNA precursor in LCLs using the ITS1–59 probe, which detects 18SE-FL, the untrimmed precursor of 18SE. (E) 18SE-FL/21S RAMP analysis of (D) and two additional biological replicates comparing BMF family member signals to the average signals of the controls 128S2, 136S1, and 157S2. (F) 18SE-FL/PTP RAMP analysis as in (E). In (B), (C), (D), p-values were calculated using one-way ANOVA of log-transformed data followed by Dunnett’s post-hoc test and adjustment of p-values for multiple comparisons. In (E) and (F) one-way ANOVA and Kruskal-Wallis test were used, respectively.
