FIGURE1. Two types of base editing.
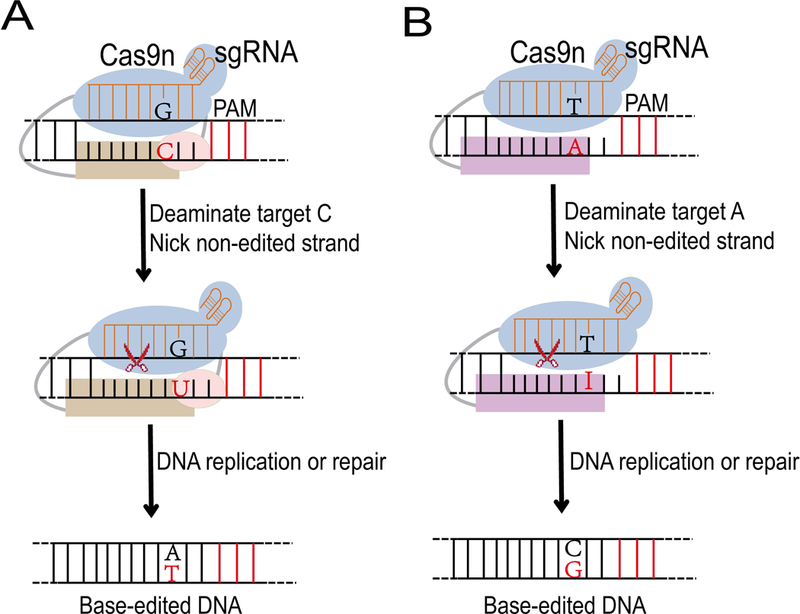
(A) Apolipoprotein B mRNA editing catalytic enzyme (APOBEC)-based cytosine base editor (CBE) mediates G•C to A•T base editing. Cas9 nikase (Cas9n, blue), which is guided by a single guide RNA (sgRNA, orange), targets specific cytosine (C, red), nicks the non-edited strand in genomic DNA, and mediates separation of local DNA strands. A tethered APOBEC enzyme (brown) acts on the C in the targeted DNA single-strand, and mediates it to uracil (U). A tethered uracil DNA alycosylase inhibitor (UGI, pink) can block the base excision to protect G•U intermediate. The resulting G•U heteroduplex can be permanently converted to an A•T base pair through DNA replication or DNA repair.
(B) Adenine base editor (ABE) mediated A•T to G•C base editing. Cas9n (blue), which is guided by a gRNA (orange), targets specific adenine (A, red), nicks the non-edited strand in genomic DNA, and mediates separation of local DNA strands. An engineered TadA* enzyme (purple) acts on the A in the DNA single-strand, and deaminates it to inosine (I). The resulting T•I heteroduplex can be permanently converted to a C•G base pair through DNA replication or DNA repair. PAM: protospacer-adjacent motif.
