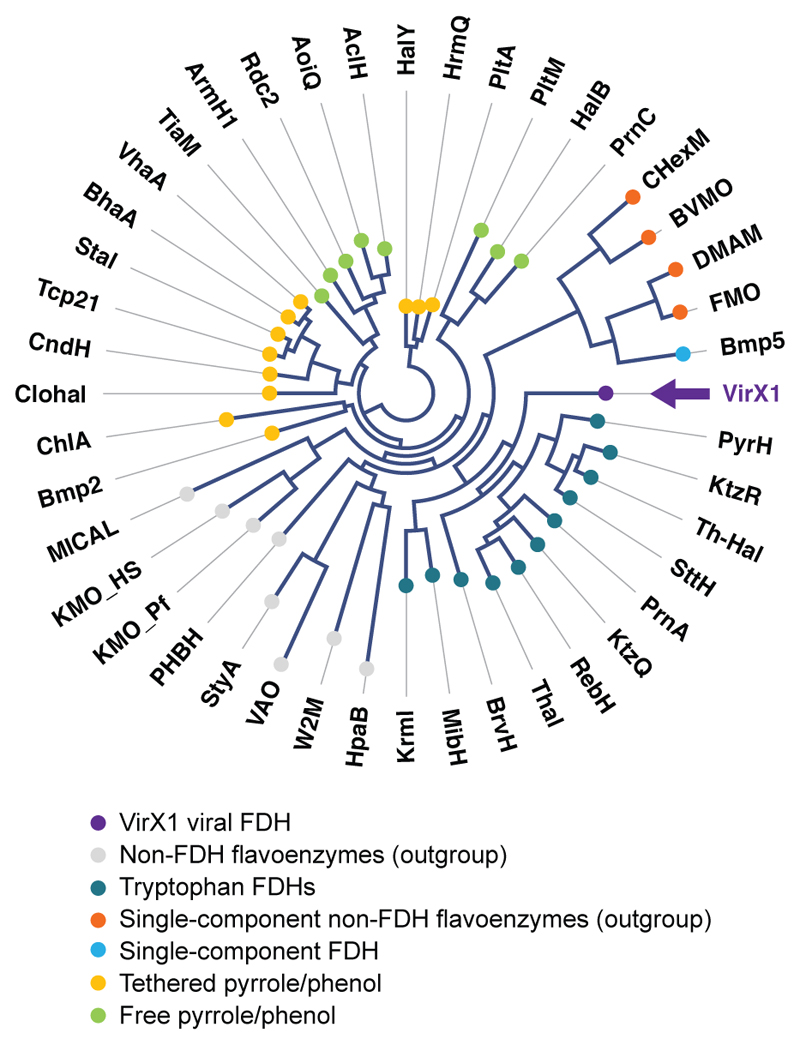
Fig. 1. Protein based branching analysis of VirX1 against known FDHs and other non-FDH flavoenzymes, revealing functional relationship, known substrate utilisation and branching order between protein clusters.
VirX1, the viral iodinase (highlighted in purple with arrow), is predicted to branch away from the tryptophan chlorinases and brominases, while still being associated with their cluster. Analysis highlights the evolved differences in halide preference and correspondingly grouped substrate preferences of these FDHs. The diagram is represented as an unrooted circular phylogram in which nodes (shown as coloured circles) represent each individual amino acid sequence analysed and branches (the lines connecting them) represent the degree of evolutionary divergence (e.g. number of amino acid substitutions, deletions or insertions that have occurred between connected branch points). A global alignment of the full-length amino acid sequences of VirX1 with other known FDHs and non-FDH flavoenzymes (used as an outgroup) was used to construct the branching diagram (through application of neighbor joining method) applying the Kimura algorithm for protein distance measurements. Bootstrapping analysis was performed in 100 replicates.