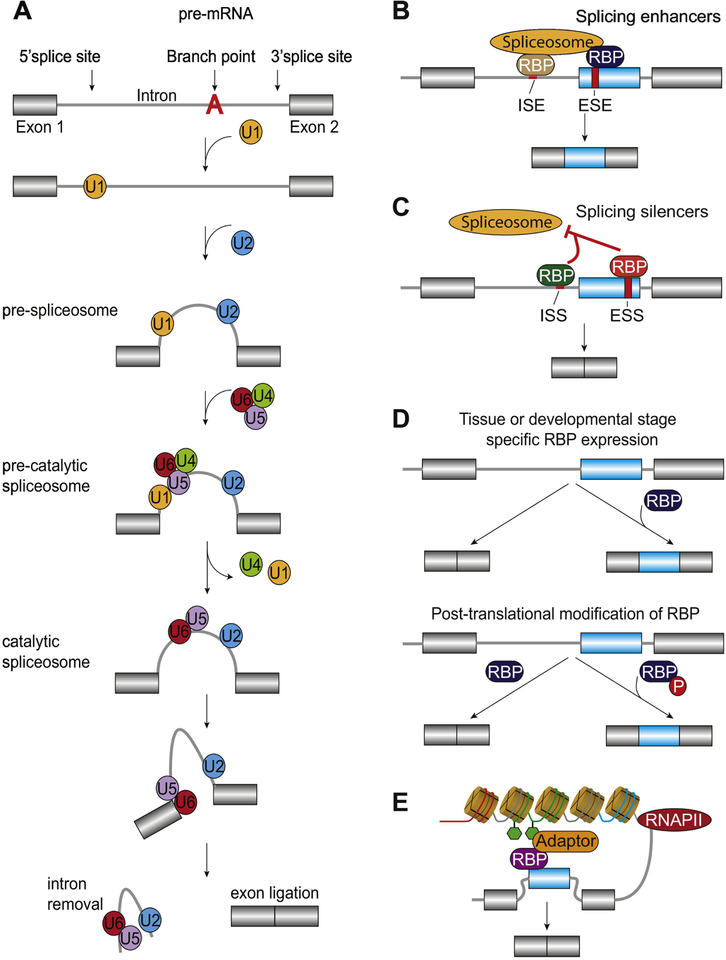
Figure 1.
Regulation of constitutive and alternative splicing. A. Assembly of the spliceosome begins with the recognition of the 5’splice site by the U1 snRNP. The pre-spliceosome complex is formed by recognition of U2 snRNP onto the branch point sequence. The pre-catalytic spliceosome is created by recruitment of the complex formed of snRNP of U4, U5 and U6. The catalytic spliceosome is completed through dissociation of U4 and U1. The final step is to excise the intronic regions and ligate remaining exonic sequences through transesterification reactions. Adapted from Shi [154]. B-C. Binding of RNA binding proteins (RBPs) to intronic or exonic splicing enhancers (ISE and ESE, respectively) or silencers (ISS and ESS, respectively) promotes exon retention (B) and skipping (C), respectively [155]. D. RBP expression is regulated in a tissue and developmental specific manner and post-translational modifications such as phosphorylation (shown as P in red) impact their ability to recognize cis-regulatory elements [155]. E. Alternative splicing is coupled to RNA polymerase II (RNAPII) kinetics and epigenetic modifications through adaptor proteins that concurrently recognize histone modifications and RBPs. Adapted from Luco et al. [6].