Figure 2.
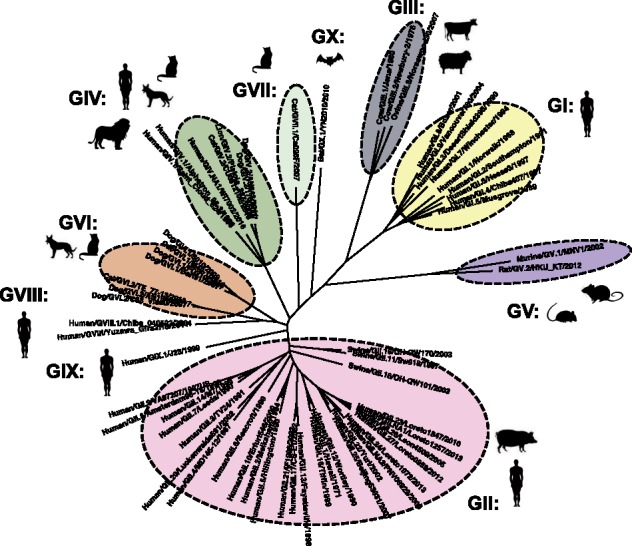
Classification of noroviruses based on the phylogeny of the major capsid protein (VP1). Genogroups are based on phylogenetic clustering and amino acid differences. Genogroups can be further divided into genotypes. Each genogroup is indicated with different colors and associated with infection of specific species (indicated by shadow figures). Viruses from genotypes GII.11, GII.18, and GII.19 infect porcine species, and viruses from GIV.1 and GIV.NA1 infect humans. Abbreviation for species: Bo, bovine; Ca, canine; Fe, feline; Hu, human; Mu, murine; Ov, ovine; Sw, swine. The phylogenetic tree was constructed using representative strains from each genotype and/or genogroup and the neighbor-joining method as implemented in MEGA (Kumar, Stecher, and Tamura 2016).
