Fig. 8.
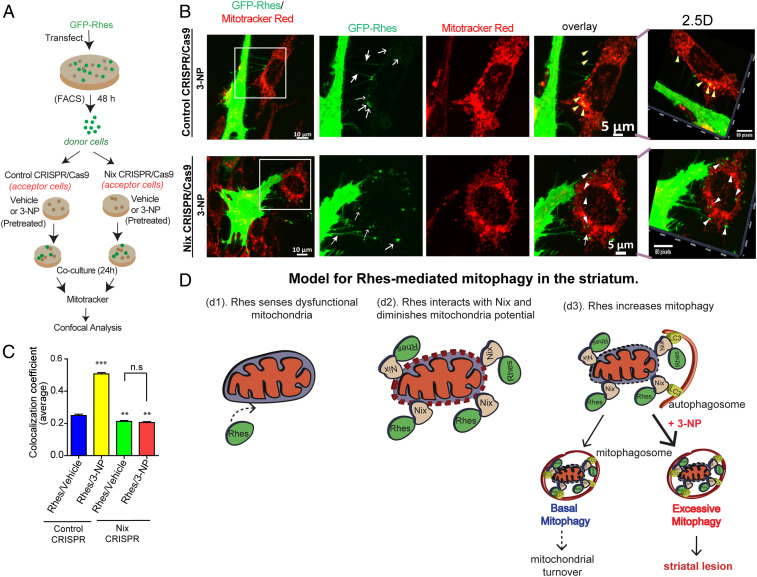
Rhes travels Intercellularly and interacts with damaged mitochondria via Nix. (A) Experimental design for B. (B) Representative confocal image of GFP-Rhes (FAC-sorted) striatal neuronal cells (donor cells) cocultured with vehicle-treated or 3-NP−treated control or Nix-depleted (Nix CRISPR) cells (acceptor cells). Insets show the magnified region from selected area. Corresponding Insets were processed for 2.5-dimensional rendering. Closed arrow indicates Rhes-induced TNT-like protrusions. Open arrow represents GFP-Rhes puncta in acceptor cell. Yellow and white arrowheads indicate presence or lack, respectively, of colocalization of GFP-Rhes with mitotracker. (C) Bar graph shows average of Pearson’s coefficient of colocalization between GFP puncta and mitotracker in neighboring acceptor cells where Rhes is transported from donor cell; n = 72 to 90 GFP-Rhes puncta per group were counted from 13 to 18 cells per group. **P < 0.01, ***P < 0.001; 1-way ANOVA followed by Tukey post hoc test; n.s, not significant. (D) Model for Rhes-mediated mitophagy in the striatum. Rhes senses dysfunctional mitochondria (d1). Rhes interacts with Nix and diminishes membrane potential (d2). Rhes-Nix interaction recruits autophagosomes via LC3 and formation of mitophagosomes to promote mitochondrial degradation (d3). Basal mitophagy may lead to mitochondrial turnover; however, excessive mitophagy in the presence of mitochondrial toxin, 3-NP, may promote striatal lesion.