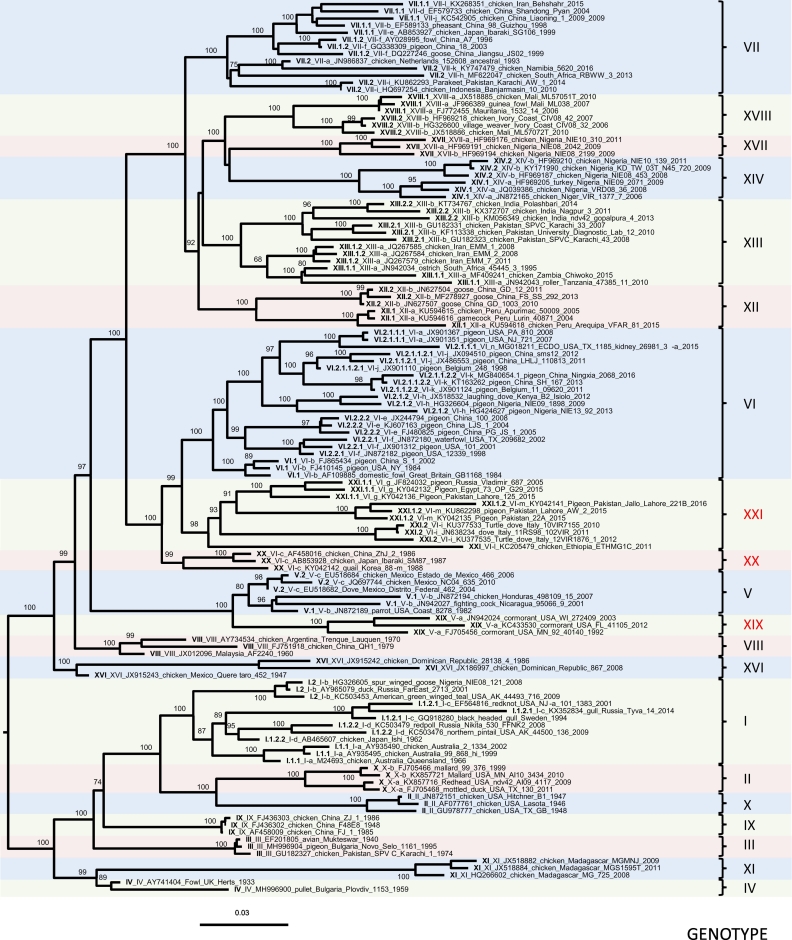
Fig. 2.
Class II “pilot” tree.
Phylogenetic analysis is based on the full-length nucleotide sequence of the fusion gene of selected isolates representing all class II Newcastle disease virus sub/genotypes (n = 125). The evolutionary history was inferred by using RaxML (Stamatakis, 2014) and utilizing the Maximum Likelihood method based on the General Time Reversible model with 1000 bootstrap replicates. The tree with the highest log likelihood (−28,785.59) is shown. A discrete gamma distribution was used to model evolutionary rate differences among sites and the rate variation model allowed for some sites to be evolutionarily invariable. The Roman numerals presented in the taxa names in the phylogenetic tree represent the respective genotype for each isolate. The new (decimal naming) and the old names (alpha-numerical) are provided for easier comparison. The taxa names also include the GenBank identification number, host name, country of isolation, strain designation, and year of isolation. Three new genotypes assigned in the current study are highlighted in red font. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)