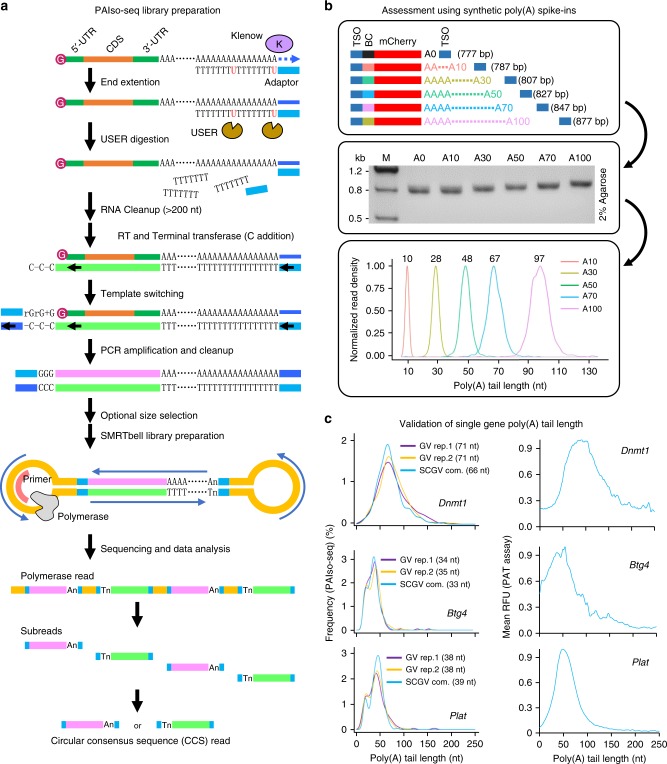
Fig. 1.
The principle and validation of PAIso−seq. a Flowchart for PAIso−seq method design. The main steps of the method include end-extension, template-switching, full-length cDNA amplification, circular adaptor ligation, and PacBio sequencing. b The structure (top panel) and agarose gel analysis of the poly(A) spike-ins (middle panel). The mean poly(A) tail length of each spike-in measured by PAIso−seq (bottom panel). Source data are provided as a Source Data file. c The poly(A) tail lengths of Dnmt1, Btg4, and Plat in GV oocytes measured by PAIso−seq (density plot of poly(A) tail length of detected CCS reads of the given genes, three replicates, left panel) and PAT assay by using capillary electrophoresis on fragment analyzer (mean of three replicates, right panel). The mean length of each gene poly(A) tail measured by PAIso−seq is shown. The number of CCS reads used are 141 (Dnmt1/GV rep.1), 249 (Dnmt1/GV rep.2), and 165 (Dnmt1/SCGV com.); 164 (Btg4/GV rep.1), 521 (Btg4/GV rep.2), and 357 (Btg4/SCGV com.); 136 (Plat/GV rep.1), 277 (Plat/GV rep.2), and 207 (Plat/SCGV com.). The average length of poly(A) tails are 74 nt (Dnmt1), 44 nt (Btg4), and 45 nt (Plat) measured by the PAT assay. RFU, relative fluorescence units.