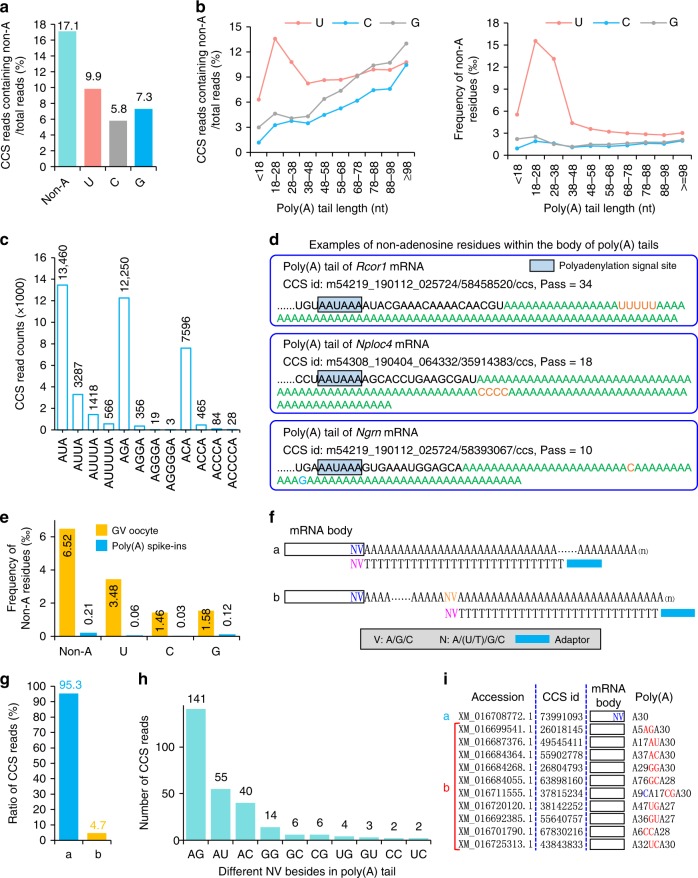
Fig. 5.
Widespread non-adenosine residues within the body of poly(A) tails. a Frequency of CCS reads containing internal non-A nucleotides within poly(A) tail. b The ratio of CCS reads containing internal non-A nucleotides (left panel) and frequency of non-adenosine residues in CCS reads of different poly(A) tail length (right panel). c Mono- and oligo-non-adenosine residue (U, C, and G) counts. d Three examples of CCS reads with non-adenosine residues in the body of poly(A) tails, Rcor1 (Pass = 34), Nploc4 (Pass = 18), and Ngrn (Pass = 10). e The frequency of non-A residues in GV oocyte and poly(A) spike-in data. f Hypothetic models of CCS reads with T30VN RT primer anchored at the end of 3′-UTR (a) or within the body of poly(A) tails (b). g Percentage of two different T30VN RT primer-anchoring models as revealed by CCS reads from Iso-seq data. h The frequency of different NV-anchoring sites detected within the body of poly(A) tails. The number of the detected events are shown above each bar. i Examples of CCS reads with T30VN RT primer anchored at the end of 3′-UTR (a) or within the body of poly(A) tails (b). The accession number of the CCS reads is shown on the left. The CCS read i.d. is shown in the middle. The CCS read model is shown on the right. The number after A means oligo A with the given number of adenosines.