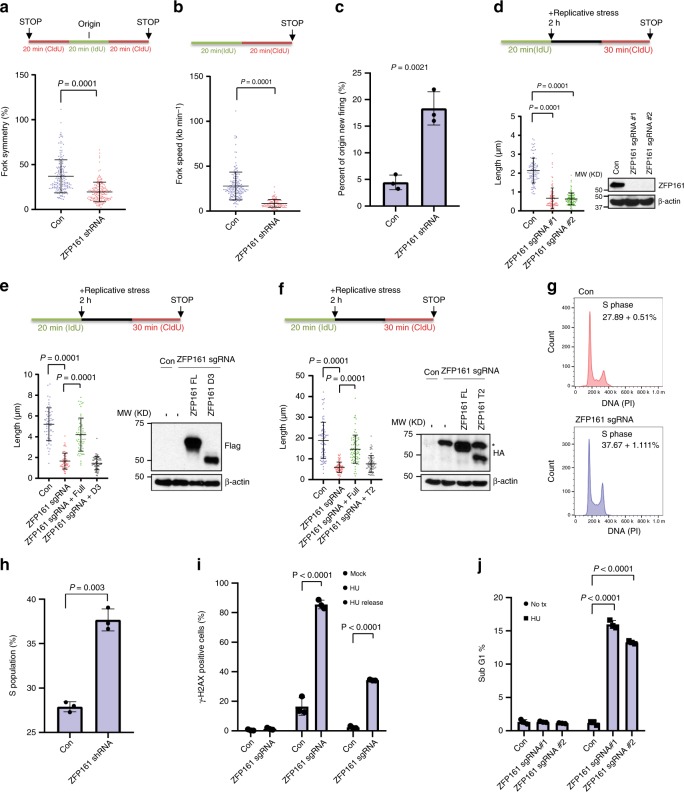
Fig. 5.
ZFP161 promotes stalled replication fork restart a, b HCT116 cells were labeled with IdU and CldU for indicated time. Fork symmetry a and fork speed b were determined by measuring the length of CIdU (red) track. c The percentage of new origins (red only fibers) was quantified. n = 313 fibers for WT and n = 285 fibers for ZFP161 knockout. d–f Cells were labeled with IdU for 20 min and then cells were incubated with 4 mM HU for 2 h. After incubation, cells were labeled with CldU for 30 min. ZFP161 KO cells (d) or KO cells reconstituted with full length ZFP161 and the D3 mutant (e), or the T2 mutant (f) were used. DNA fibers were stretched on a microscope slide, stained with IdU and CldU antibodies, imaged, and the lengths of fiber tracks measured. The graphs represent mean ± S.D., two-tailed, unpaired t-test. g, h Cell cycle distribution was determined by flow cytometry in cells either lacking or expressing ZFP161 (g). S phase populations were analyzed by Modifit (h). i, j HCT116 cells were treated with 2 mM HU for 24 h and released. Cells were collected and γ-H2AX (i) and sub G1 population (an indicator of cell death) (j) were determined by flow cytometry. *: unspecific bands The graphs represent mean ± S.D., two-tailed, paired t-test; n = 3 independent experiments. Source data are provided as a Source Data file.