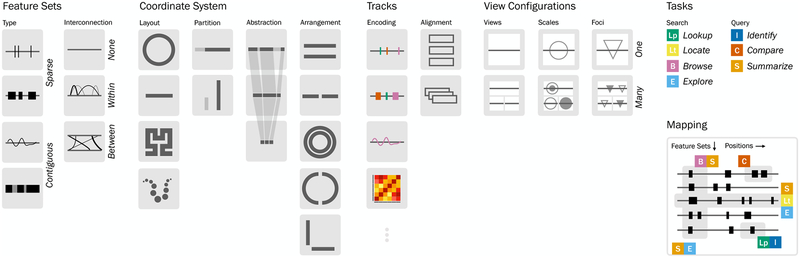
Figure 1:
Data taxonomy, visualization taxonomy, and task taxonomy for genomic visualizations. The data taxonomy describes how different genomic data types can be encoded as feature sets. A genomic visualization contains one or multiple coordinate systems applying a specific layout, partition, abstraction and arrangement (in case of multiple axes) of sequence coordinates. Feature sets are encoded as tracks and placed on the coordinate systems. Multiple tracks can either be aligned by stacking or overlaying. A visualization can consist of one or multiple views, each containing a set of aligned tracks. Multiple views can show data on one or multiple scales and foci. The task taxonomy explains how different search and query tasks operate on genomic visualizations.