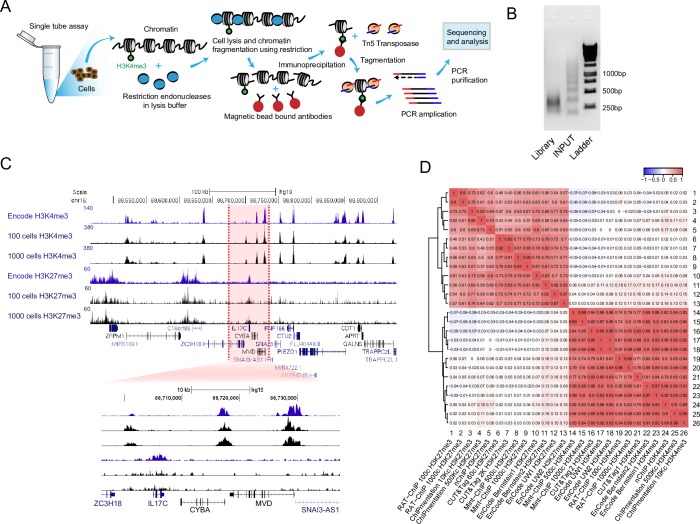
Fig 1. RAT-ChIP enables genome wide histone modification profiling from 100 cells.
A Overview of RAT-ChIP method. B Agarose gel electrophoresis of DNA after chromatin treatment with a combination of restriction enzymes (middle lane) and after tagmentation (left lane). C UCSC genome browser custom histone H3K4me3 and H3K27me3 tracks of RAT-ChIP-seq with 100 and 1,000 K562 cells in comparison with ENCODE data in a genomic region centred around IL17C gene. D Clustered global Pearson correlation heatmap (enrichments in 5kb windows) of RAT-ChIP-seq and different published histone H3K4me3 and H3K27me3 datasets in K562 cells.