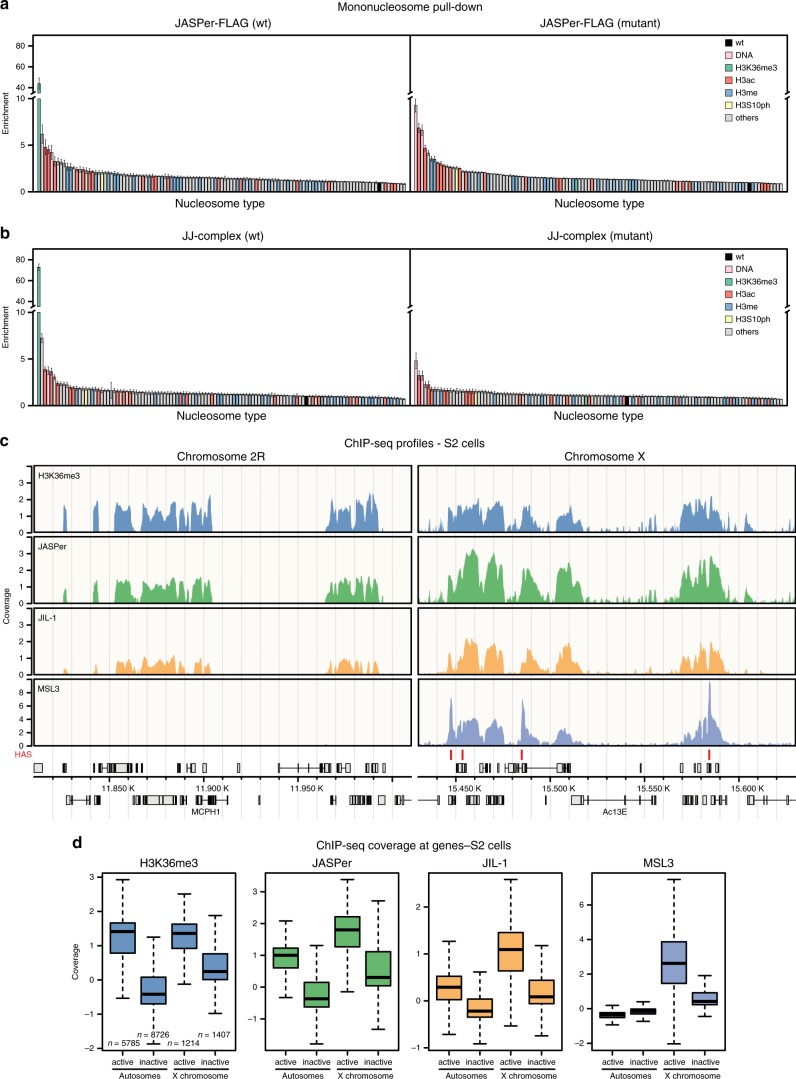
Fig. 3.
The JJ-complex binds H3K36me3 nucleosomes in vitro and in vivo, and is enriched on the male X chromosome. a Bar chart of mean enrichment (n = 3 independent experiments with 2 different protein preparations) of nucleosome library pull-down with JASPer-FLAG (left panel) and aromatic cage mutant (right panel) relative to unmodified nucleosome, which is set to 1. Error bars represent standard error of the mean. b Bar chart of mean enrichment (n = 3 independent experiments) of nucleosome library pull-down with JJ-complex (FLAG-JIL-1 and untagged JASPer) (left panel) and aromatic cage mutant (right panel) relative to unmodified nucleosome, which is set to 1, as in a. Error bars represent standard error of the mean. c Genome browser profile showing mean H3K36me3 (upper panel, n = 4 independent experiments), JASPer (second upper panel, n = 4 independent experiments with 2 different antibodies), JIL-1 (second lower panel, n = 5 independent experiments with 2 different antibodies) and MSL3 (lower panel, n = 3 independent experiments) MNase ChIP-seq normalized coverage along representative 200 kb windows on chromosome 2 R and X in male S2 cells. HAS are marked by red bars above the gene models in gray. d Box plot showing mean H3K36me3 (left panel, n = 4), JASPer (second left panel, n = 4), JIL-1 (second right, n = 5) and MSL3 (right, n = 3) MNase ChIP-seq normalized coverage, as in c, at active (tpm > 1) and inactive (tpm ≤ 1) genes on the autosomes (n = 5785 and n = 8726, respectively) and X chromosome (n = 1214 and n = 1407, respectively) in male S2 cells. Box plot elements are defined as center line marking the median, box limits are the upper and lower quartiles, whiskers extend maximally 1.5-times the interquartile range and outliers are removed.