Fig. 7.
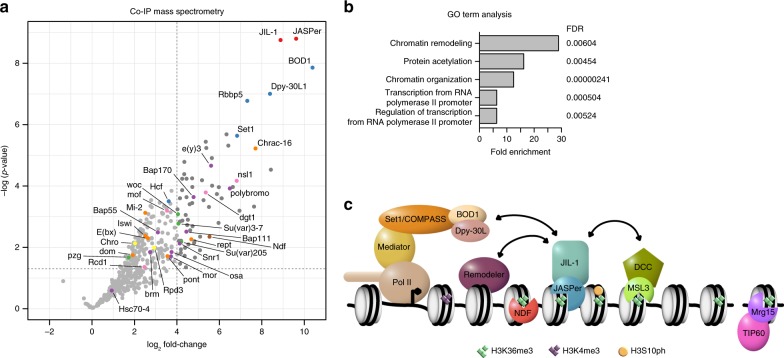
The JASPer interaction network and other H3K36me3 binding proteins and complexes in Drosophila melanogaster. a Volcano plot of IP-MS showing –log10(p-values) against mean log2 fold-change in α-JASPer IP (n = 6) versus control IP (n = 5). Significantly enriched (p-value < 0.05 and log2 fold-change > 4) proteins (n = 69) are highlighted in dark gray. JIL-1 and JASPer are marked in red, Set1/COMPASS complex members in blue, PBAP/Brm complex members in purple, other proteins involved in chromatin remodeling in orange, NSL complex members in pink, Su(var)3-7, Su(var)205, woc and pzg in green, Chro and Rpd3 in yellow and NDF in brown. b Bar plot showing GO term enrichment of significantly enriched proteins shown in a. The five statistically significantly (fdr < 0.01) most enriched GO terms are shown. c Model of JJ-complex binding at H3K36me3 marked gene bodies and interactions with other complexes. Interactions presented here are indicated by arrows. Other known H3K36me3 binding proteins (NDF and Mrg15) are drawn at the lower side. We propose that phosphorylation by JIL-1 kinase is tightly regulated in space and time in part by its partner JASPer which stabilizes and anchores the kinase to active genes and telomeric transposons by binding to H3K36me3 nucleosomes via its PWWP domain. Regulation by JASPer affects any potential phosphorylation by JIL-1, in particular H3Ser10 phosphorylation which is involved in inhibition of heterochromatinisation.