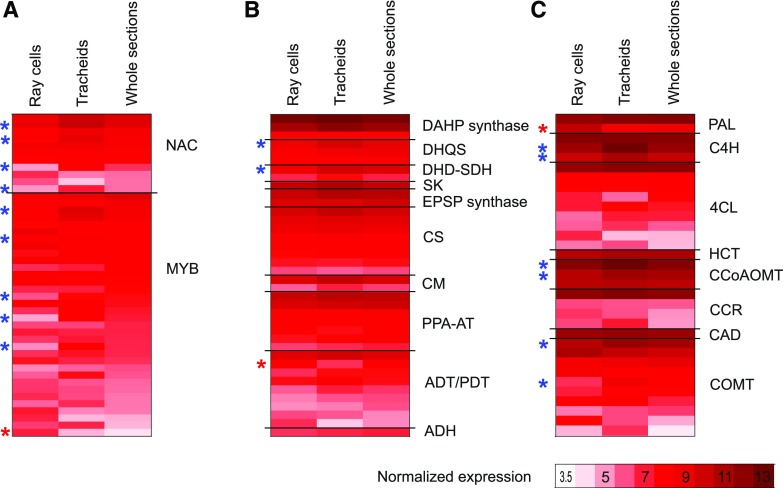
Figure 4.
Average-normalized expression of genes important in lignification. NAC and MYB transcription factors (A), shikimate, Phe, and Tyr pathway genes leading to Phe and Tyr (B), and phenylpropanoid and monolignol biosynthesis pathway genes leading to monolignols (C) are shown in ray parenchymal cells, upright tracheids, and whole sections of developing xylem with expression levels higher than 5 (variance-stabilizing transformation [VST] > 5) at least in one of the sample types. Red stars count for significantly higher and blue for significantly lower levels of expression in ray cells than in tracheids (Padj ≤ 0.05). Shikimate, Phe, and Tyr pathway genes are according to Maeda and Dudareva (2012): ADH, arogenate dehydrogenase; ADT, arogenate dehydratase; CM, chorismate mutase; CS, chorismate synthase; DAHP synthase, 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase; DHD-SDH, 3-dehydroquinate dehydratase-shikimate dehydrogenase; DHQS, 3-dehydroquinate synthase; EPSP synthase, 5-enolpyruvylshikimate 3-phosphate synthase; PDT, prephenate dehydratase; PPA-AT, prephenate aminotransferase; SK, shikimate kinase. General phenylpropanoid pathway genes are as follows: CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase; PAL, phenylalanine ammonia lyase. Monolignol pathway genes are as follows: CAD, cinnamyl alcohol dehydrogenase; CCR, cinnamoyl-CoA reductase; COMT, caffeate/5-hydroxyconiferaldehyde O-methyltransferase. p-Coumaroyl shikimate 3-hydroxylase is missing from the figure due to annotation difficulties related to cytochrome P450 genes, as enzyme activity cannot be inferred from the gene sequence. See Supplemental Table S3 for gene identifiers.