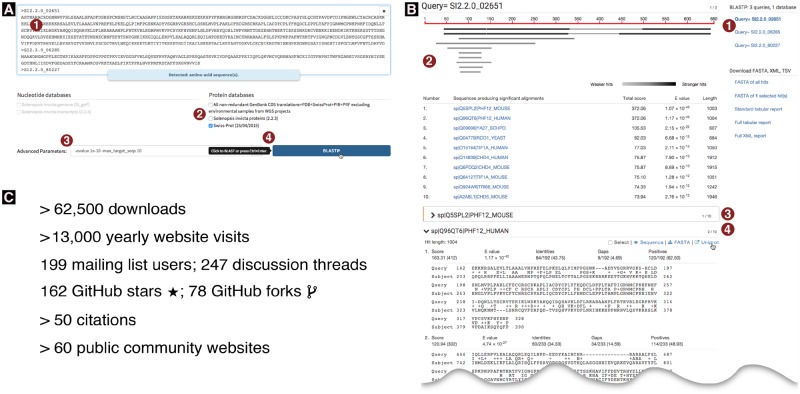
Fig. 1.
(A) Partial screenshot of the query interface. Numbers circled in red highlight the steps involved and some specific features. (1) Three or more sequences were pasted into the query field (typewriter font; only the identifier is visible for the third sequence); a message confirms to the user that these are amino acid sequences. (2) The Swiss-Prot protein database was the first database to be selected. As a result, additional database selections are limited to protein databases; nucleotide databases are disabled. (3) Optional advanced parameters were entered which constrain the results to the ten strongest hits with E-values stronger than 10−10. (4) The BLAST button is automatically activated and labeled “BlastP” as this is the only possible basic BLAST algorithm for the given query-database combination. As the user’s mouse pointer hovers over the BlastP button, a tooltip indicates that a keyboard shortcut exists for this button. (B) Partial screenshot of a Sequenceserver BLAST report. An interactive version of this figure is online at http://sequenceserver.com/paper/resultsinteractive (last accessed August 25, 2019). Three amino acid sequences were compared against the Swiss-Prot database using BlastP with an E-value cutoff of 10−10 and keeping only the ten strongest hits per query. This screenshot shows a portion of the results for the first query. Numbers circled in red highlight some specific features of this report. (1) An index overview summarizes the query and database information and provides clickable links to query-specific results. (2) Results for the first query are shown. These include a graphical overview indicating which parts of the query sequence align to each hit, a tabular summary of all hits, and alignment details for each hit. (3) The first hit is selected for download; its alignment details have been folded away. (4) The user is studying the second hit; the mouse pointer hovers over the link to the hit’s UniProt page. (C) Sequenceserver usage as of June 11, 2019. These include download statistics from https://rubygems.org/gems/sequenceserver, Google Analytics statistics for http://sequenceserver.com, and citation statistics from https://app.dimensions.ai/details/publication/pub.1085102830, and GitHub statistics from https://github.com/wurmlab/sequenceserver.