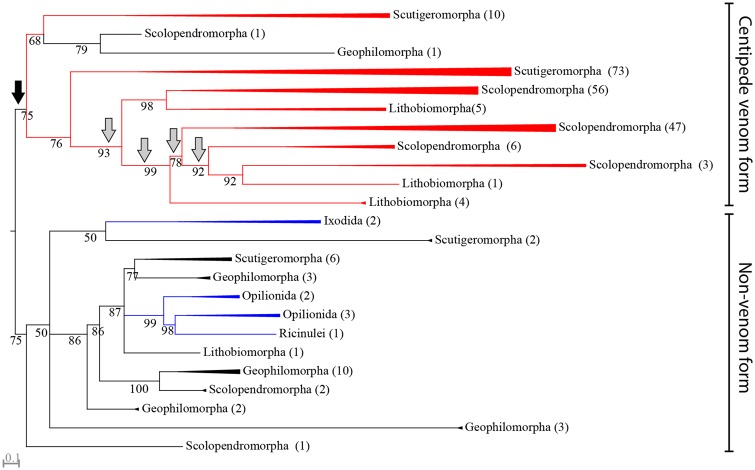
Fig. 4.
Gene duplication and loss drives the evolution of β-PFTx venom proteins. Maximum likelihood (ML) phylogenetic reconstruction of the β-PFTx family (under WAG+F+R5, chosen according to Bayesian Information Criterion) displayed as rooted with a clade containing taxonomic outgroup sequences. Gene tree/species tree reconciliation suggests a single gene duplication occurred in the ancestral centipede, indicated by a black arrow, whereas four subsequent duplications occurred along the stem lineage of Pleurostigmophora, indicated by gray arrows. Functional losses were associated with all these suggested duplication events. Clades containing only sequences from a single order are collapsed, with the number of sequences in each collapsed clade shown in parentheses. Clades containing sequences identified in venom proteomes are colored red, whereas noncentipede sequences are colored blue. Bootstrap support values are shown at each node, and nodes with support <50 are collapsed into polytomies. For the phylogenetic tree without collapsed clades see supplementary materialfigure S3 (Supplementary Material online).