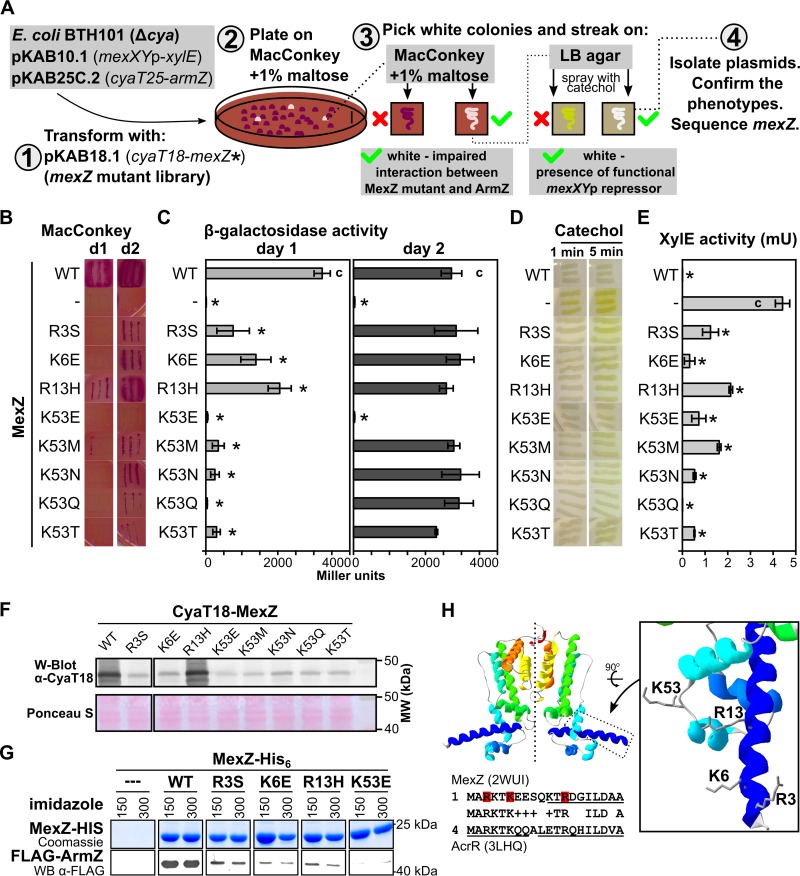
FIG 2.
Positively charged amino acids in the N-terminal part of MexZ are engaged in interaction with ArmZ. (A) Overview of the strategy used for identification of MexZ mutants with disturbed interactions with ArmZ but that retained the ability to repress mexXYp. (B) BACTH analysis of the interactions between MexZ single-substitution variants and ArmZ. Transformants carrying pKAB10.1, pKAB25C.2, and plasmids expressing cyaT18-mexZ (WT or the indicated mutants) were restreaked onto selective MacConkey plates and grown for 1 day (d1) or 2 days (d2). (C) β-galactosidase activity in cultures of transformants from panel B. (D) Ability of MexZ mutants to repress mexXYp-xylE. Transformants (as in panels B and C) were grown on L agar, sprayed with 10 mM catechol, and photographed after 1 and 5 min. White color indicates repression of mexXYp-xylE by MexZ. (E) XylE activity assayed in extracts from exponentially growing strains as described above for panel B. β-Galactosidase and XylE activity data represent mean values from at least three assays ± SD. *, P < 0.05 (as determined by two-sided Student’s t test) relative to controls (indicated with “c”). (F) Assessment of CyaT18-MexZ levels by Western blotting in cultures used for XylE activity assays. Anti-CyaT18 antibodies were used, and a Ponceau S-stained membrane is shown as a loading control. (G) Pulldown analysis of WT and mutant MexZ interactions with FLAG-ArmZ. A total of 150 μg of purified MexZ-His6 (WT or mutants) and 1 mg of proteins from an extract of E. coli cells overproducing FLAG-ArmZ were applied to Ni-NTA columns, followed by washing and elution using an imidazole gradient. Proteins were separated by SDS-PAGE, followed by Coomassie staining or Western blot analysis using anti-FLAG antibodies. For clarity, the images were cropped and the contrast was enhanced (original images are included in Fig. S2 in the supplemental material). (H) Amino acids participating in the MexZ-ArmZ interaction visualized on the refined MexZ structural model based on data reported previously (29). At the bottom is an alignment of the N-terminal part of MexZ and S. Typhimurium AcrR (42). Residues identified in this study as being involved in interactions with ArmZ are indicated in red. Alpha helices detected in crystal structures are underlined.