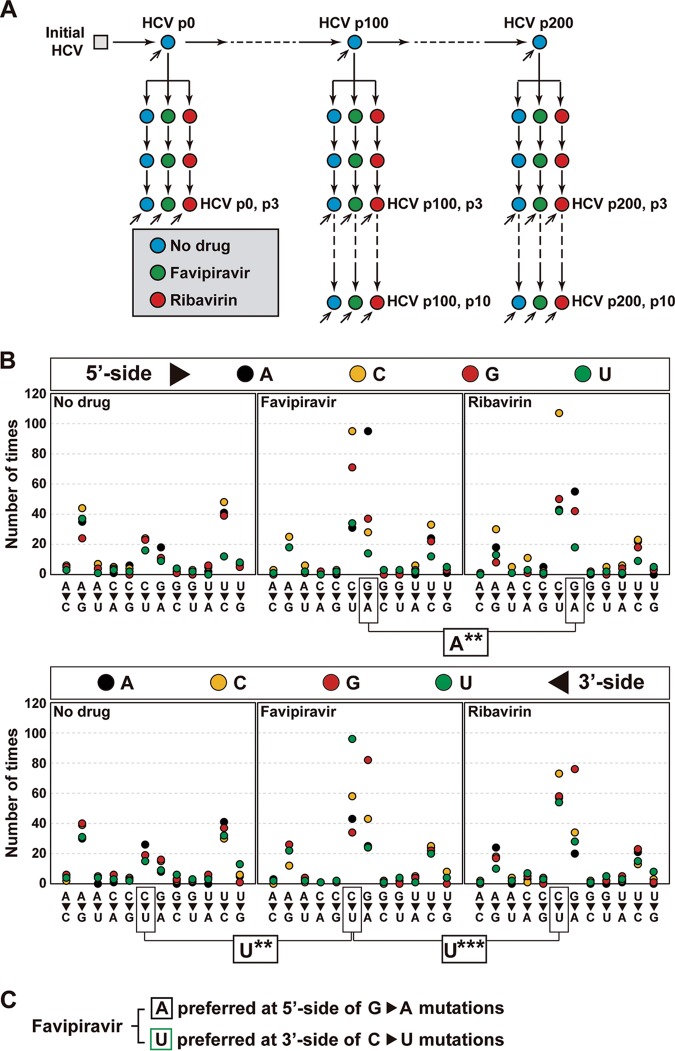
FIG 3.
Influence of the immediate neighbors of each mutated site on mutant frequency. (A) Scheme of HCV passages in the absence (No drug) and the presence of favipiravir or ribavirin. Inclined arrows indicate the viral population analyzed by MiSeq Illumina deep sequencing of three amplicons of the NS5B-coding region. (B) Effect of the nucleotide type present at the 5′ and 3′ sides of the mutated residue. The mutation types are given on the abscissa, and the number of times that a given neighbor is present is given on the ordinate (the color code for the nucleotide is in the box at the top). Statistical significances (determined by the proportion test) are given below each panel group, with boxes linking the relevant comparisons. **, P < 0.01; ***, P < 0.001. Deep sequencing, data processing, and statistical procedures are described in Materials and Methods. (C) Summary of statistically significant preferred neighbor nucleotides for mutation sites.