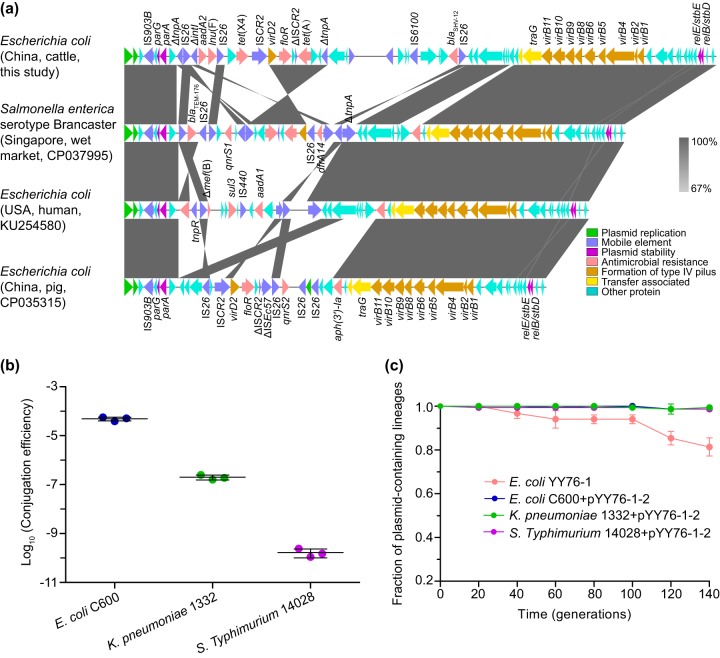
FIG 1.
(a) Comparative analysis of IncX1 plasmids. The arrows represent the positions and transcriptional directions of the open reading frames. Regions of homology ranging from 67% to 100% are marked by shading. The Δ symbol indicates that the gene is truncated. (b) Conjugation transfer efficiencies of the tet(X4)-harboring IncX1 plasmid pYY76-1-2 into E. coli, K. pneumoniae, and S. Typhimurium strains. The transfer efficiency (mean ± standard deviation) is calculated based on colony counts of the transconjugant and recipient cells in triplicate. (c) Plasmid stability experiment results. The fraction of plasmid-containing cells (mean ± standard deviation) is plotted against the passage generations in triplicate.