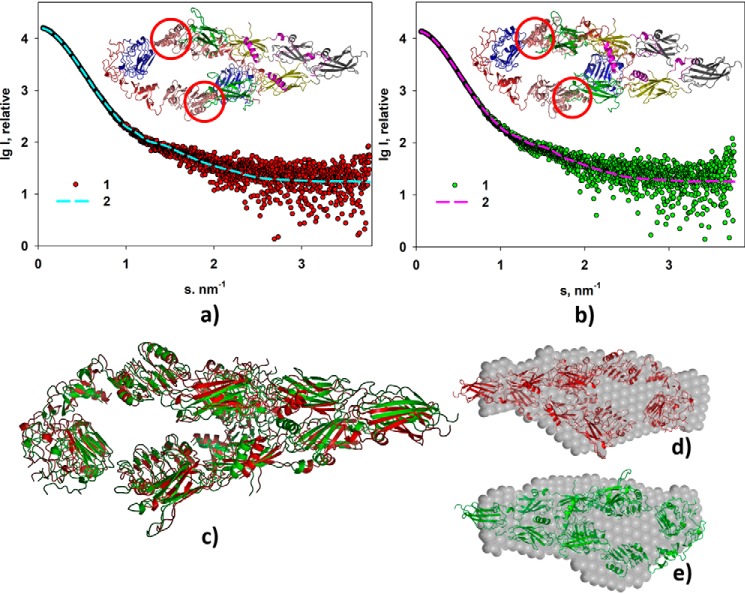
Figure 3.
Rigid body modeling of the ectoIRR dimer structure. a, modeling at pH 7. 0. b, modeling at pH 9.0. Experimental scattering curves (1) and scattering patterns computed from the CORAL models (2) are shown. Insets, structural organizations of the ectoIRR dimer in solution obtained by CORAL. Red circles mark obtained configuration of amino acids 466–471 as a dummy-residue chain to fit the computed scattering patterns. c, comparison of the CORAL models obtained for pH 7.0 (red) and for pH 9.0 (green); NSD = 0.97. d and e, comparison of the CORAL models with the corresponding DAMMIN shapes at pH 7.0 and 9.0, respectively; NSD = 1.3. P1 symmetry was used for all shape reconstructions.