Abstract
Several enzymes of the CYP716A subfamily have been reported to be involved in triterpenoid biosynthesis. Members of this subfamily oxidize various positions along the triterpenoid backbone and the majority of them catalyze a three-step oxidation at the C-28 position. Interestingly, C-28 oxidation is a common feature in oleanolic acid, ursolic acid, and betulinic acid, which are widely distributed in plants and exhibit important biological activities. In this work, three additional CYP716A enzymes isolated from olive, sugar beet, and coffee, were characterized as multifunctional C-28 oxidases. Semi-quantitative comparisons of in vivo catalytic activity were made against the previously characterized enzymes CYP716A12, CYP716A15, and CYP716A52v2. When heterologously expressed in yeast, the isolated enzymes differed in both catalytic activity and substrate specificity. This study indicates that the screening of enzymes from different plants could be a useful means of identifying enzymes with enhanced catalytic activity and desired substrate specificity. Furthermore, we show that “naturally-evolved” enzymes can be useful in the heterologous production of pharmacologically and industrially important triterpenoids.
Keywords: Beta vulgaris, Coffea arabica, CYP716A, Olea europaea, triterpenoid
Introduction
Oleanolic, ursolic, and betulinic acids are important triterpenoids with diverse biological activities, including antifungal, antibacterial, anti-HIV, and/or antitumor activities (Fulda and Kroemer 2009; Perez 2003; Sasazuka et al. 1995; Wu 2009). These compounds are spread widely among plants and can be found in olive, coffee, and sugar beet (Fai and Tao 2009).
The fruits of the olive tree (Olea europaea L., Oleaceae), a subtropical evergreen native to the Mediterranean coast, are consumed worldwide as food. Olive wax contains multiple triterpenoids, predominantly uvaol, erythrodiol, ursolic, and oleanolic acids. In addition, whole fruits also contain β-amyrin and betulinic acid (Bianchi et al. 1992). Oleanolic acid, ursolic acid, maslinic acid, uvaol, and erythrodiol are also found in olive oil as minor constituents (Allouche et al. 2009).
Sugar beet (Beta vulgaris, Chenopodiaceae) is a hardy biennial plant, originating from around the Mediterranean Sea (Doney 1986), which accounts for approximately 16% of the world’s sugar production (FAO 2012). Oleanolic acid, a reportedly active participant in hepatoprotection (Yabuchi et al. 1988), and seco-glycosides thereof have been isolated from sugar beet leaves and roots (Massiot et al. 1994).
Coffea arabica (coffee, Rubiaceae), a woody perennial plant cultivated originally in the Arabian peninsula, accounts for 60% of the world’s coffee production (ITC 2011). Ursolic acid and oleanolic acid have been detected in mature coffee leaves (Waller et al. 1991).
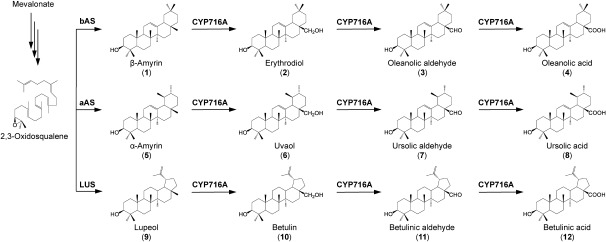
The biosynthesis of oleanolic, ursolic, and betulinic acids (Figure 1) is initiated with the cyclization of 2,3-oxidosqualene to a triterpenol, namely β-amyrin, α-amyrin, and lupeol, respectively. Each of these reactions is catalyzed by a specific oxidosqualene cyclase (OSC): β-amyrin synthase, α-amyrin synthase, and lupeol synthase, respectively (Herrera et al. 1998; Iturbe-Ormaetxe et al. 2003; Kushiro et al. 1998; Morita et al. 2000; Suzuki et al. 2002). Subsequently, cytochrome P450 enzymes modify these triterpenols, yielding the corresponding C-28 oxidized products. Recently, several C-28 oxidizing enzymes have been identified in various plant species (Carelli et al. 2011; Fukushima et al. 2011; Miettinen et al. 2017; Seki et al. 2015). The industrial production of C-28 oxidized products is an active research area (Li and Zhang 2015; Zhou et al. 2016). Interestingly, the yields of oxidized products were dependent on both the expression system and the origin of the expressed enzymes. Moreover, the catalytic activity of P450 is considered a limiting factor for the accumulation of C-28 oxidized products (Czarnotta et al. 2017).
Figure 1. Biosynthetic pathway of C-28 oxidized triterpenoids. Three step oxidations catalyzed by CYP716A enzymes at the C-28 position of three triterpene skeletons are shown. bAS, β-amyrin synthase; aAS, α-amyrin synthase; LUS, lupeol synthase.
In this study, we identified three CYP716A enzymes, obtained from olive, sugar beet, and coffee, as multifunctional oxidases in the biosynthesis of oleanolic, ursolic, and betulinic acids. We report a semi-quantitative comparison of the in vivo catalytic activities of these enzymes, expressed heterologously in yeast, against those of the previously characterized Medicago truncatula CYP716A12, Vitis vinifera CYP716A15, and Panax ginseng CYP716A52v2 enzymes.
Methods
Cloning and plasmid construction
CYP716A48 (Olea europaea)
A partial sequence of CYP716A48 (accession no. G0244188.1) was retrieved from expressed sequence tags (ESTs) deposited in the National Center for Biotechnology Information (NCBI) (OEAA-070810_Plate3p13.b1 cDNA library from olive leaves and fruits, O. europaea cDNA, mRNA sequence). The C-terminal region was deduced from homology-based comparisons. A recent search of the olive genome database (http://denovo.cnag.cat/genomes/olive/) yielded four homologous genes showing at least 85% amino acid identity.
Total RNA was prepared from the leaves of O. europaea (cv. Nevadilo Blanco) plants using RNeasy (Qiagen, Hilden, Germany) and cDNA synthesis was conducted using a GeneRacer™ Kit (Invitrogen, Carlsbad, CA, USA). Primers 1–2 (Supplementary Table S1) were used to amplify the full-length open reading frame, which was then cloned into pENTR™/D-Topo® to generate pENTR-CYP716A48.
CYP716A49 (Beta vulgaris)
The N-terminal region (TC8082, Beet Release 2.0, May 19, 2008) was retrieved from the Gene Index Project Site (http://compbio.dfci.harvard.edu/tgi=tgipage.html) of the Dana-Farber Cancer Institute, Harvard School of Public Health Gene Index Project (http://compbio.dfci.harvard.edu/tgi/tgipage.html). The C-terminal region (accession no. FG44256.1) was obtained from the NCBI BLAST site (SF_01-a06 Bv_T1 B. vulgaris ssp. vulgaris cDNA clone SF_01-a06 reverse, mRNA sequence). CYP716A49 exhibited 99.1% amino acid identity with B. vulgaris CYP716A (partial) published in the Cytochrome P450 Homepage (Nelson 2009). A recent search of the sugar beet genome database (http://bvseq.molgen.mpg.de/index.shtml) yielded a single homologous gene showing 73% amino acid identity with CYP716A49.
Total RNA was prepared from one-week-old seedlings of B. vulgaris (cv. Detroit Dark Red) plants using RNeasy (Qiagen) and the SuperScriptTM III first-strand synthesis system (Invitrogen). Primers 3–4 (Supplementary Table S1) were used to amplify the full-length sequence. The sequence was then transferred to pENTR™/D-Topo® to generate pENTR-CYP716A49.
CYP716A50 (Coffea arabica)
A partial sequence of CYP716A50 was retrieved from ESTs deposited in the NCBI database (accession no. GT020247.1, TransId-279 278 CarcatBudEnri2 C. arabica cDNA clone CarCatBudEnri2_59-D09-PAL17d similar to cytochrome P450 monooxygenase CYP716A12—M. truncatula mRNA sequence). The C-terminal region was deduced by homology-based comparisons.
Total RNA was prepared from frozen leaves of C. arabica (cv. Blue mountain) plants using RNeasy (Qiagen) and cDNA synthesis was conducted using a GeneRacer™ Kit (Invitrogen). Primer 5, together with primer 5′ from the kit, was used for the first amplification and primers 6–7 (Supplementary Table S1) were used to amplify the full-length sequence. The sequence was then transferred to pENTR™/D-Topo® to generate pENTR-CYP716A50.
CYP716A52v2 (Panax ginseng)
P. ginseng has two CYP716A52 variants (v1 and v2), the latter of which has been characterized by Hang et al. (2013). Total RNA was prepared from P. ginseng callus cells (a gift from Nitto Denko Corporation) using TRIzol™ reagent (Ambion, Carlsbad, CA, USA) and an RNeasy Plant Mini Kit (Qiagen). cDNA synthesis was performed using PrimeScript™ RT Master Mix (Perfect Real Time) (Takara, Tokyo, Japan). Primers 8–9 (Supplementary Table S1) were used to amplify the full-length sequence. The sequence was then transferred to pENTR™/D-Topo® to generate pENTR-CYP716A52v2.
Plasmid construction
The pYES3-ADH-OSC1 plasmid (for constitutive expression of Lotus japonicus bAS under the control of the ADH1 promoter) and pELC-GW (for galactose-inducible expression of L. japonicus CPR alone) were constructed as described previously (Seki et al. 2008). Similarly, pYES3-ADH-aAS and pYES3-ADH-LUS (for constitutive expression of O. europaea aAS and Glycyrrhiza uralensis LUS respectively, both under the control of the ADH1 promoter) constructs have been described previously (Fukushima et al. 2011).
The coding sequences (CDSs) of CYP716A48, CYP716A49, CYP716A50, and CYP716A52v2 were transferred into pELC-GW and pYES-DEST52 (Thermo Fisher Scientific, Waltham, MA, USA) vectors using GW LR Clonase II Enzyme Mix (Thermo Fisher Scientific) to create pELC-CYP716A48, pELC-CYP716A49, pELC-CYP716A50, pELC-CYP716A52v2, pDEST52-CYP716A48, pDEST52-CYP716A49, pDEST52-CYP716A50, and pDEST52-CYP716A52v2.
Yeast in vivo assays
Saccharomyces cerevisiae INVSc1 (MATa his3D1 leu2 trp1-289 ura3-52; Invitrogen) harboring pYES3-ADH-aAS, pYES3-ADH-OSC1, or pYES3-ADH-LUS was co-transformed with pELC-CYP716A and pDEST52-CYP716A using Frozen-EZ Yeast Transformation II™ (Zymo Research, Irvine, CA, USA). Recombinant yeast cells were cultured in synthetic complete medium (5 ml) containing 2% glucose without tryptophan, leucine, and uracil (SC-W-L-U) until an OD600 of 1.3–1.7 at 30°C and 180 rpm was reached. The cells were collected, resuspended in SC-W-L-U medium (10 ml) with 2% galactose to induce CYP716As expression, and cultured overnight at 30°C and 180 rpm. The obtained cultured samples were stored at −30°C until use. All assays were performed in triplicate (three independent assays obtained from different clones). Samples harboring pYES3-ADH-aAS, pYES3-ADH-OSC1, or pYES3-ADH-LUS together with the empty vectors pELC-GW and pYES-DEST52 were used as controls.
Extraction and GC–MS analysis
Authentic hederagenin was added as an internal standard prior to extraction. Yeast cultures were extracted thrice with 6 ml of ethyl acetate (Wako, Osaka, Japan), followed by 30 s of vortexing and 30 min of sonication, and concentrated by centrifugal evaporation. The remaining powder was transferred to a silica-gel column (6 cc; Waters Corp., Milford, MA, USA) and washed with 10 ml ethyl acetate. The samples were then placed in an evaporator until dry. The obtained pellet was resuspended in 1 ml (bAS- and aAS-expressing lines) or 400 µl (LUS-expressing lines) of 1 : 1 chloroform–methanol solution and 100 µl was transferred to a vial and placed in an evaporator until dry. Finally, the pellet was derivatized with 50 µl of N-methyl-N-(trimethyl silyl) trifluoro acetamide (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at 80°C. The evaporated samples were stored at −30°C until use.
Gas chromatography–mass spectrometry (GC–MS) analyses were performed using a 5977 A MSD mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) connected to a 7890B gas chromatograph (Agilent Technologies) with a DB-1 ms (30 m×0.25 mm, 0.25 µm film thickness; Agilent Technologies) capillary column. The injection temperature was 250°C. The column temperature program was as follows: 80°C for 1 min, an increase to 260°C at a rate of 20°C/min, an increase to 280°C at a rate of 10°C/min and hold for 20 min, and a final increase to 320°C at a rate of 20°C/min and hold for 16 min. The carrier gas was helium at a flow rate of 1.0 ml/min. The ion source temperature was 230°C and the quadrupole temperature was 150°C with a splitless injection. Detected peaks were identified by comparing the retention times (Rts) and mass spectra with those of authentic standards.
Extracted ion chromatograms based on the following m/z values were used for calculating peak areas and product yields: 218 (β-amyrin), 216 (erythrodiol), 203 (oleanolic acid, α-amyrin, ursolic acid, betulin, and betulinic acid), 496 (uvaol), 189 (lupeol), and 320 (hederagenin).
Chemicals (sapogenin authentic standards)
All purchased sapogenins were at least of analytical grade. β-Amyrin (≥98.5% purity), α-amyrin (≥98.5% purity), lupeol (≥99% purity), erythrodiol (≥97% purity), uvaol (≥98.5% purity), oleanolic acid (≥99% purity), ursolic acid (≥98.5% purity), and hederagenin (≥98.5% purity) were purchased from Extra Synthese (Lyon, France). Betulin (≥98% purity) was purchased from Sigma-Aldrich. Betulinic acid (≥97% purity) was purchased from Tokyo Chemical Industry (Tokyo, Japan). Oleanolic aldehyde, ursolic aldehyde, and betulinic aldehyde were synthesized as indicated in Supplementary Data S1.
Phylogenetic tree analyses
Full-length amino acid sequences were collected from GenBank and aligned with MUSCLE (Edgar 2004). A phylogenetic tree was generated using the neighbor joining method with 1,000 replicates using MEGA6 software (Tamura et al. 2013). Protein sequences are listed in Supplementary Data S2.
Results
Functional analysis of CYP716As
To examine the abilities of CYP716A48, CYP716A49, and CYP716A50 to convert simple triterpenoid substrates, we heterologously co-expressed CYP716A48, CYP716A49, and CYP716A50 with CPR and β-amyrin synthase, α-amyrin synthase, or lupeol synthase, respectively (Fukushima et al. 2011; Fukushima et al. 2014). For comparison, the previously characterized enzymes CYP716A12, CYP716A15, and CYP716A52v2 (Fukushima et al. 2011; Han et al. 2013), were also tested.
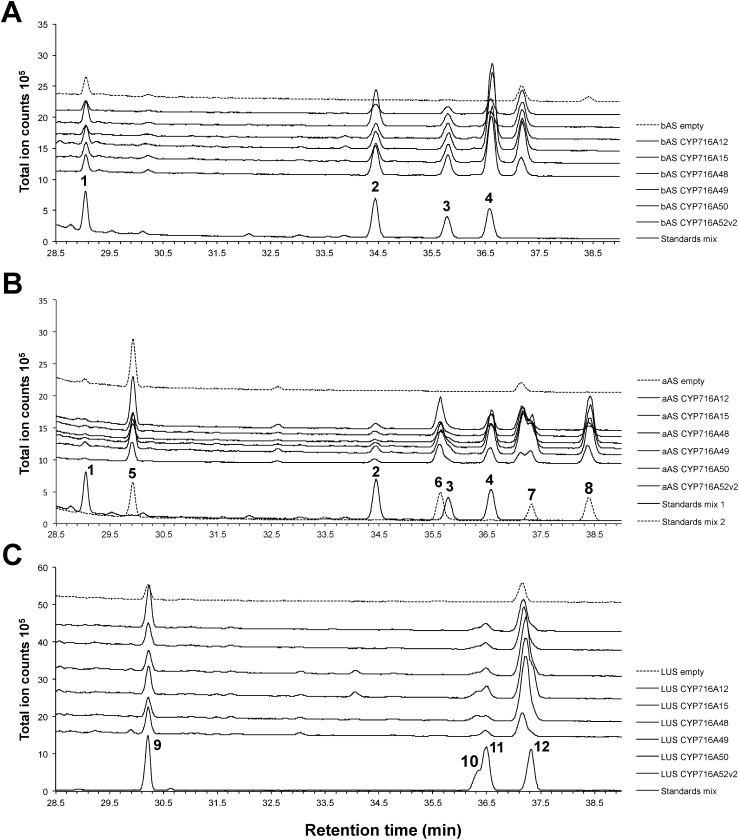
The total ion chromatogram (TIC) of an ethyl acetate extract of bAS/CPR/CYP716As-expressing yeast contained three peaks that were not present in the TIC of the control sample (carrying bAS and CPR alone) (Figure 2A). Peak 2 (Rt=34.44 min) was identified as erythrodiol (compound 2 in Figure 1), peak 3 (Rt=35.79 min) was identified as oleanolic aldehyde (compound 3 in Figure 1), and peak 4 (Rt=36.57 min) was identified as oleanolic acid (compound 4 in Figure 1) based on comparisons with Rts and mass spectra of authentic standards (Supplementary Figure S1).
Figure 2. In vivo activity of CYP716A enzymes. Total ion chromatograms (TICs) of ethyl acetate extracts from (A) bAS/CPR/CYP716A-expressing yeast, (B) aAS/CPR/CYP716A-expressing yeast, and (C) LUS/CPR/CYP716A-expressing yeast are shown. Peak numbers shown in the chromatograms correspond to those in Figure 1.
Peaks corresponding to uvaol (6; Rt=35.63 min), ursolic aldehyde (7; Rt=37.32 min), and ursolic acid (8; Rt=38.40 min) were found in the TIC of an ethyl acetate extract of aAS/CPR/CYP716As-expressing yeast (Figure 2B). The amyrin synthase we introduced to generate α-amyrin-producing yeast has been shown to also produce β-amyrin (Saimaru et al. 2007). We therefore detected erythrodiol (2), oleanolic aldehyde (3), and oleanolic acid (4) as minor products. Finally, peaks corresponding to betulin (10; Rt=36.33 min), betulinic aldehyde (11; Rt=36.46 min), and betulinic acid (12; Rt=37.29 min) were found in an ethyl acetate extract of LUS/CPR/CYP716As-expressing yeast (Figure 2C). None of the above-mentioned peaks were observed in the control samples.
Together, these results confirm that, when heterologously expressed in transgenic yeast, CYP716A48, CYP716A49, and CYP716A50 are able to modify β-amyrin (1), α-amyrin (5), and lupeol (9) to produce oleanolic acid (4), ursolic acid (8), and betulinic acid (12), respectively. Similarly, CYP716A52v2, in addition to its ability to produce oleanolic acid (Han et al. 2013), is able to produce ursolic and betulinic acids.
Semi-quantitative analyses
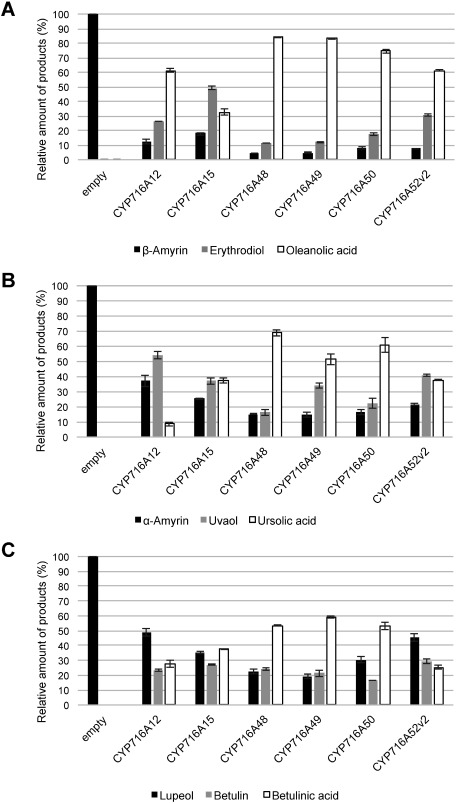
Employing a semi-quantitative method with an internal standard, we compared the accumulation of C-28 oxidized triterpenoids resulting from the heterologous expression of the above-characterized CYP716As in transgenic yeast (Figure 3). The amounts of simple triterpenoids in control samples were considered to be 100%. In CYP716As-expressing samples, the sum of simple triterpenoids and C-28 hydroxylated and carboxylated derivatives were considered to be 100%. In general, C-28 aldehyde compounds were excluded due to the instability of authentic standards. The exception was betulinic aldehyde (11), which co-eluted with betulin (10) (Figure 2). Peak 10 in Figure 3 corresponds to the sum of both compounds.
Figure 3. Relative quantification of C-28 oxidized triterpenoids. The relative amounts of (A) β-amyrin and its derivatives in bAS/CPR/CYP716A-expressing yeast extracts, (B) α-amyrin and its derivatives in aAS/CPR/CYP716A-expressing yeast extracts, and (C) lupeol and its derivatives in LUS/CPR/CYP716A-expressing yeast extracts, are shown. Quantitation and error bars correspond to the mean and standard deviation, respectively (n=3).
Each of the tested enzymes showed a strong oxidation activity for the conversion of simple triterpenoids into carboxylic acids. However, differences were observed in both substrate preference and catalytic efficiency.
The most preferred substrate, as indicated by the highest conversion efficiencies, was β-amyrin. The percent remaining substrate ranged from 4.4% with CYP716A48 to 18.1% with CYP716A15. With the exception of bAS/CYP716A15-expressing yeast, in which hydroxylated product (49.2%) accumulated in greater amounts than its carboxylated derivative (32.7%), other bAS/CYP716A-expressing yeasts yielded accumulations of oleanolic acid higher than those of erythrodiol (from 61.2% with CYP716A12 to 84% with CYP716A48). The second-most preferred substrate was α-amyrin, with 15% and 37.1% unconverted substrate with CYP716A49 and CYP716A12, respectively. Although the conversion rate was sufficient to accumulate ursolic acid (Figure 3 and Supplementary Figure S2), the yield of ursolic acid was three- to five-fold lower than that of oleanolic acid. Similarly, the least-preferred substrate, lupeol, was 19.3% and 49% unconverted by CYP716A49 and CYP716A12, respectively. While it could be converted to betulinic acid, the yields were six- to seventeen-fold lower than that of oleanolic acid (Figure 3 and Supplementary Figure S2).
Catalytic efficiency was also reflected in the yields of oxidized compounds. The yield of oleanolic acid ranged from 273.2±57.7 µg l−1 (CYP716A15) to 1953.5±394.8 µg l−1 (CYP716A49) in bAS/CPR/CYP716A-expressing yeasts. Ursolic acid production ranged from 62.2±10.3 µg l−1 (CYP716A12) to 360.2±124.8 µg l−1 (CYP716A48) in aAS/CPR/CYP716A-expressing yeasts, and the amount of betulinic acid ranged from 37.6±7.0 µg l−1 (CYP716A12) to 104.6±33.7 µg l−1 (CYP716A49) in LUS/CPR/CYP716A-expressing yeasts (Supplementary Figure S2).
Discussion
We previously characterized three members of the CYP716A subfamily found in M. truncatula and V. vinifera as β-amyrin-(1), α-amyrin-(5), and lupeol-(9) C-28 oxidizing enzymes involved in the biosynthesis of oleanolic, ursolic, and betulinic acids (4, 8, 12) (Fukushima et al. 2011). In this study, we report C-28 oxidizing activity in transgenic yeast for three additional subfamily CYP716A members (CYP716A48, CYP716A49, and CYP716A50). We also confirmed the C-28 oxidizing activity of the previously characterized CYP716A52v2 on lupeol (9) and α-amyrin (5). Moreover, we semi-quantitatively compared the catalytic activities and substrate specificities of the above-mentioned CYP716A enzymes when expressed in yeast as a heterologous host.
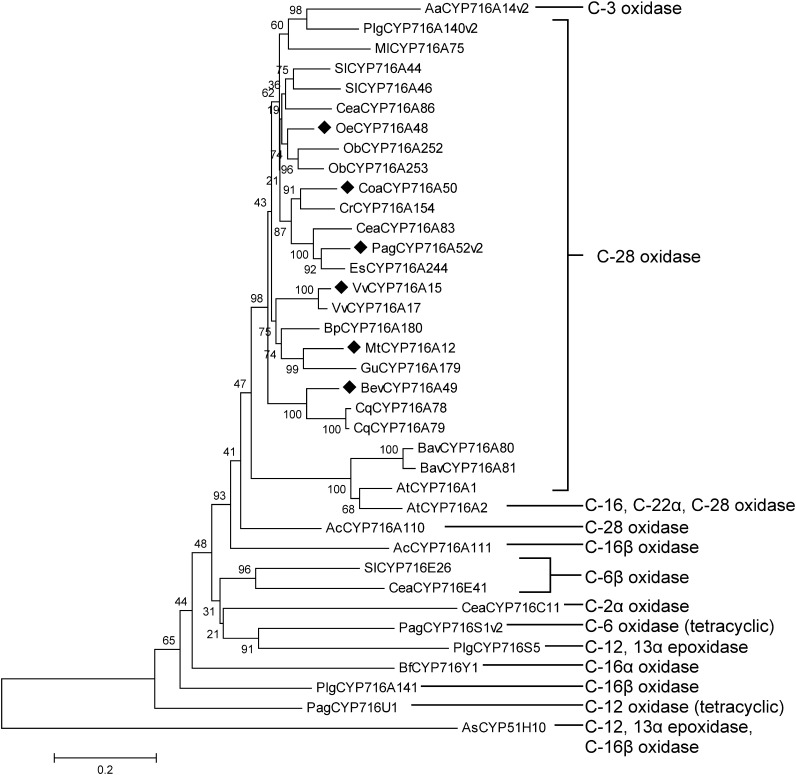
This study contributes to an increased understanding of the role of CYP716A enzymes in the diversification of the triterpenoid backbone (Figure 4; Miettinen et al. 2017; Misra et al. 2017; Tamura et al. 2017a, 2017b; Yasumoto et al. 2016, 2017). We also identified previously reported putative compounds, oleanolic, ursolic, and betulinic aldehydes (3, 7, 11), by synthesizing authentic standards and comparing their Rts and mass spectra with those of chromatographic peaks obtained from heterologous yeast extracts.
Figure 4. Phylogenetic tree of triterpene biosynthesis-related CYP716 family enzymes. The phylogenetic tree was generated as described in the Materials and methods. The scale bar corresponds to the number of amino acid substitutions per site. The activities toward triterpene skeletons are indicated on the right. The enzymes used in this study are highlighted. Aa, Artemisia annua; Plg, Platycodon grandiflorus; Ml, Maesa lanceolata; Sl, Solanum lycopersicum; Cea, Centella asiatica; Oe, Olea europaea; Ob, Ocimum basilicum; Coa, Coffea arabica; Cr, Catharanthus roseus; Pag, Panax ginseng; Es, Eleutherococcus senticosus; Vv, Vitis vinifera; Bp, Betula platyphylla; Mt, Medicago truncatula; Gu, Glycyrrhiza uralensis; Bev, Beta vulgaris; Cq, Chenopodium quinoa; Bav, Barbarea vulgaris; At, Arabidopsis thaliana; Ac, Aquilegia coerulea; Bf, Bupleurum falcatum; As, Avena strigosa.
Our findings agree with those of previous reports regarding the accumulation of C-28 oxidized triterpenoids in their corresponding plant sources, namely olive, sugar beet, and coffee (Allouche et al. 2009; Kondo et al. 1971; Massiot et al. 1994; Waller et al. 1991). O. europaea, B. vulgaris and C. arabica accumulate mainly oleanolic acid. While able to oxidize α-amyrin, β-amyrin and lupeol CYP716A48, CYP716A49 and CYP716A50 showed a preference for β-amyrin as a substrate. Similarly, M. truncatula accumulates oleanane-type triterpenoids (Huhman and Sumner 2002) and CYP716A12 mainly oxidized β-amyrin. P. ginseng accumulates dammarane- and oleanane-type triterpenoids. CYP716A52v2 has been previously characterized as a β-amyrin oxidase (Hang et al. 2013) and our results confirm that it prefers β-amyrin as a substrate. Although more detailed analyses are required, such as metabolite profiles and functional characterization in planta, our results suggest an evolutionary relationship between the catalytic activity of CYP716A enzymes and triterpenoid accumulation in the reported plants.
Although the CYP716As evaluated here belong to different plant families, namely Oleaceae, Chenopodiaceae, Rubiaceae, Araliaceae, Fabaceae, and Vitaceae, they show more than 70% identity at the amino acid level (Figure 4; Supplementary Table S2) and exhibited similar behaviors as C-28 oxidases. Our semi-quantitative analyses, however, revealed significant differences in catalytic activity and substrate specificity. These differences might arise from several factors. One may be the relative GC content of the genes, which can affect protein translation efficiency (CYP716A12, 28%; CYP716A15, 49%; CYP716A48, 43%; CYP716A49, 46%; CYP716A50, 44%; and CYP716A52v2, 47%). However, this does not adequately explain the differences encountered herein since the enzymes exhibiting the lowest catalytic activities differ widely in their GC content (CYP716A12, 28% and CYP716A15, 49%).
Differences in the amino acid sequences of the tested enzymes may account for differences in catalytic activity. However, the crystal structure of CYP716A is not currently available and any prediction of key amino acids determining catalytic activity and substrate specificity would be speculative. As shown in Supplementary Figure S3, the tested enzymes differed in several amino acid motifs in both the N- and C-terminal regions, making it difficult to precisely identify the important residues.
Many other factors, depending on the particular protein and the host organism, such as mRNA stability, transfer RNA copies, protein folding kinetics, protein stability, protein transport, and toxicity of the protein within the expression environment, may influence the expression of a gene (Cannarozzi et al. 2010; Wang et al. 2002; Zhang and Ignatova 2011). Recent findings show that protein–protein interactions in planta are important in the orchestration of metabolic pathways and efficient metabolic flux (Laursen et al. 2016). Thus, subtle differences at the amino acid level among the tested enzymes may affect not only substrate interactions, but may also affect the OSCs and CPRs co-expressed in our yeast system.
Synthetic engineering plays an important role in enhancing the production of natural products. These, in turn, play a dominant role in the discovery and development of pharmaceuticals (Newman and Cragg 2016). For C-28 oxidized triterpenoids, the main bottleneck for production in a heterologous host is the catalytic activity of P450 enzymes (Czarnotta et al. 2017). Common strategies, such as codon optimization and protein engineering, may help overcome this bottleneck, but this is not a guarantee. In this scenario, the use of “naturally-evolved” enzymes could be an alternative means of boosting the production efficiency of commercially important triterpenoids (Moses et al. 2014). We confirmed that the screening of enzymes from different plant origins could be useful for selecting those with both enhanced catalytic activity and desired substrate specificity.
Acknowledgements
This study was financially aided in part by the Frontier Research Base for Global Young Researchers, Osaka University, supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, to EOF and a Grant-in-Aid for Scientific Research on Innovative Areas, No. JP17H05442, to H.S.
Supplementary Data
References
- Allouche Y, Jiménez A, Uceda M, Aguilera MP, Gaforio JJ, Beltrán G (2009) Triterpenic content and chemometric analysis of virgin olive oils from forty olive cultivars. J Agric Food Chem 57: 3604–3610 [DOI] [PubMed] [Google Scholar]
- Bianchi G, Murelli C, Vlahov G (1992) Surface waxes from olive fruits. Phytochemistry 31: 3503–3506 [Google Scholar]
- Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y (2010) A role for codon order in translation dynamics. Cell 141: 355–367 [DOI] [PubMed] [Google Scholar]
- Carelli M, Biazzi E, Panara F, Tava A, Scaramelli L, Porceddu A, Graham N, Odoardi M, Piano E, Arcioni S, et al. (2011) Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23: 3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnotta E, Dianat M, Korf M, Granica F, Merz J, Maury J, Baallal Jacobsen SA, Förster J, Ebert BE, Blank LM (2017) Fermentation and purification strategies for the production of betulinic acid and its lupane-type precursors in Saccharomyces cerevisiae. Biotechnol Bioeng 114: 2528–2538 [DOI] [PubMed] [Google Scholar]
- Doney DL (1986) Origin, distribution and collection of sugarbeet and its wild relatives. Sugarbeet Research and Extension Reports 16: 292–294 [Google Scholar]
- Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and highthroughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fai YM, Tao CC (2009) A review of presence of oleanolic acid in natural products. Natura Proda Medica 2: 1–271 [Google Scholar]
- FAO (2012) http://www.fao.org/nr/water/cropinfo_sugarbeet.html
- Fukushima EO, Seki H, Ohyama K, Ono E, Umemoto N, Mizutani M, Saito K, Muranaka T (2011) CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol 52: 2050–2061 [DOI] [PubMed] [Google Scholar]
- Fukushima EO, Seki H, Muranaka T (2014) Heterologous expression of triterpene biosynthetic genes in yeast and subsequent metabolite identification through GC-MS. In: Manuel R. (ed) Plant Isoprenoids: Methods in Molecular Biology (Methods and Protocolos), Vol. 1153. Humana Press, pp 235–243 [DOI] [PubMed]
- Fulda S, Kroemer G (2009) Targeting mitochondrial apoptosis by betulinic acid in human caners. Drug Discov Today 14: 885–890 [DOI] [PubMed] [Google Scholar]
- Han JY, Kim MJ, Ban YW, Hwang HS, Choi YE (2013) The involvement of β-amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol 54: 2034–2046 [DOI] [PubMed] [Google Scholar]
- Herrera JBR, Bartel B, Wilson WK, Matsuda S (1998) Cloning and characterization of the Arabidopsis thaliana lupeol synthase gene. Phytochemistry 49: 1905–1911 [DOI] [PubMed] [Google Scholar]
- Huhman DV, Sumner LW (2002) Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59: 347–360 [DOI] [PubMed] [Google Scholar]
- International Trade Center (ITC) (2011) The Coffee Guide: World production by type: Arabica and robusta. http://www.thecoffeeguide.org//coffee-guide/world-coffee-trade/world-production-by-type-arabica-and-robusta/
- Iturbe-Ormaetxe I, Haralamidis K, Papadopoulou K, Osbourn AE (2003) Molecular cloning and characterization of triterpene synthases from Medicago truncatula and Lotus japonicus. Plant Mol Biol 51: 731–743 [DOI] [PubMed] [Google Scholar]
- Kondo N, Marumoto Y, Shoji I (1971) Studies on the constituents of Panacis japonica rhizoma. IV. The structure of chikusetsusaponin V. Chem Pharm Bull 19: 1103–1107 [Google Scholar]
- Kushiro T, Shibuya M, Ebizuka Y (1998) β-amyrin synthase: Cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur J Biochem 256: 238–244 [DOI] [PubMed] [Google Scholar]
- Laursen T, Borch J, Knudsen C, Bavishi K, Torta F, Martens HJ, Silvestro D, Hatzakis N, Wenk M, Dafforn T, et al. (2016) Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science 354: 890–893 [DOI] [PubMed] [Google Scholar]
- Li J, Zhang Y (2015) Modulating betulinic acid production in Saccharomyces cerevisiae by managing the intracellular supplies of the co-factor NADPH and oxygen. J Biosci Bioeng 119: 77–81 [DOI] [PubMed] [Google Scholar]
- Massiot G, Dijoux MG, Lavaud C, Le Men-Olivier L, Connolly JD, Sheeley DM (1994) Seco-glycosides of oleanolic acid from Beta vulgaris. Phytochemistry 37: 1667–1670 [DOI] [PubMed] [Google Scholar]
- Miettinen K, Pollier J, Buyst D, Arendt P, Csuk R, Sommerwerk S, Moses T, Mertens J, Sonawane PD, Pauwels L, et al. (2017) The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis. Nat Commun 8: 14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra RC, Sharma S, Sandeep AG, Chanotiya CS, Ghosh S (2017) Two CYP716A subfamily cytochrome P450 monooxygenases of sweet basil play similar but nonredundant roles in ursane- and oleanane-type pentacyclic triterpene biosynthesis. New Phytol 214: 706–720 [DOI] [PubMed] [Google Scholar]
- Morita M, Shibuya M, Kushiro T, Masuda K, Ebizuka Y (2000) Molecular cloning and functional expression of triterpene synthases from pea (Pisum sativum): New α-amyrin-producing enzyme is a multifunctional triterpene synthase. Eur J Biochem 267: 3453–3460 [DOI] [PubMed] [Google Scholar]
- Moses T, Thevelein JM, Goossens A, Pollier J (2014) Comparative analysis of CYP93E proteins for improved microbial synthesis of plant triterpenoids. Phytochemistry 108: 47–56 [DOI] [PubMed] [Google Scholar]
- Nelson DR (2009) The cytochrome P450 homepage. Hum Genomics 4: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79: 629–661 [DOI] [PubMed] [Google Scholar]
- Perez RM (2003) Antiviral activity of compounds isolated from plants. Pharm Biol 41: 107–157 [Google Scholar]
- Saimaru H, Orihara Y, Tansakul P, Kang YH, Shibuya M, Ebizuka Y (2007) Production of triterpene acids by cell suspension cultures of Olea europaea. Chem Pharm Bull 55: 784–788 [DOI] [PubMed] [Google Scholar]
- Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Tamura K, Muranaka T (2015) P450s and UGTs: Key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol 56: 1463–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Achnine L, Xu R, Matsuda SP, Dixon RA (2002) A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J 32: 1033–1048 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Seki H, Suzuki H, Kojoma M, Saito K, Muranaka T (2017a) CYP716A179 functions as a triterpene C-28 oxidase in tissue-cultured stolons of Glycyrrhiza uralensis. Plant Cell Rep 36: 437–445 [DOI] [PubMed] [Google Scholar]
- Tamura K, Teranishi Y, Ueda S, Suzuki H, Kawano N, Yoshimatsu K, Saito K, Kawahara N, Muranaka T, Seki H (2017b) Cytochrome P450 monooxygenase CYP716A141 is a unique β-amyrin C-16β oxidase involved in triterpenoid saponin biosynthesis in Platycodon grandiflorus. Plant Cell Physiol 58: 874–884 [DOI] [PubMed] [Google Scholar]
- Waller GR, Jurzysta M, Karns TKB, Geno PW (1991) Isolation and identification of ursolic acid from Coffea arabica L. (coffee) leaves. In: Proceedings of the 14th International Scientific Colloquium on Coffee, San Francisco–USA. www.asic-cafe.org/pdf/abstract/14_028.pdf
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO (2002) Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA 99: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CD (2009) Grape products and oral health. J Nutr 139: 1818S–1823S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuchi T, Tanaka T, Sasatsuka T, Yamahara J, Fujimura H (1988) Extraction of oleanolic acid from sugar beets for treatment of liver failure. Chem Abstr 108: 82082 [Google Scholar]
- Yasumoto S, Fukushima EO, Seki H, Muranaka T (2016) Novel triterpene oxidizing activity of Arabidopsis thaliana CYP716A subfamily enzymes. FEBS Lett 590: 533–540 [DOI] [PubMed] [Google Scholar]
- Yasumoto S, Seki H, Shimizu Y, Fukushima EO, Muranaka T (2017) Functional characterization of CYP716 family P450 enzymes in triterpenoid biosynthesis in tomato. Front Plant Sci 8: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ignatova Z (2011) Folding at the birth of the nascent chain: Coordinating translation with co-translational folding. Curr Opin Struct Biol 21: 25–31 [DOI] [PubMed] [Google Scholar]
- Zhou C, Li J, Li C, Zhang Y (2016) Improvement of betulinic acid biosynthesis in yeast employing multiple strategies. BMC Biotechnol 16: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.