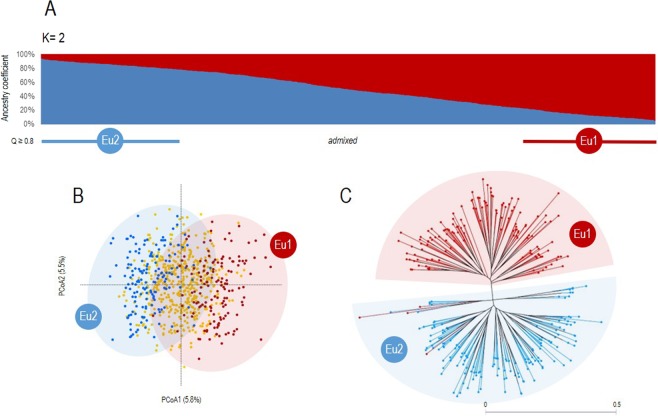
Figure 1.
Population structure of grape phylloxera in Europe. In A, population structure results obtained with STRUCTURE39 considering 774 grape phylloxera MLGs and 7 SSR loci is shown. The optimal number of genetic groups (K = 2, Eu1 and Eu2) was set considering the method of Puechmaille40. Every MLG is shown as a vertical line, whose colour(s) indicates their estimated membership to Eu1 (red) or Eu2 (blue). Considering a critical ancestry coefficient of Q ≥ 0.80, 143 and 159 MLGs were assigned to Eu1 and Eu2, respectively (472 MLGs were considered as admixed). In B, a principal coordinate analysis (PCoA) obtained from a dissimilarity matrix calculated in DARwin from genetic data (7 SSRs) from 752 MLGs is shown. MLGs assigned to Eu1 and Eu2 are indicated as red and blue dots, respectively. Admixed MLGs are shown as yellow dots. The variance explained by the PCoA1 and PcoA2 is indicated (%). In C, the neighbor-joining unweighted dendrogram obtained in DARwin considering the MLGs grouped in Eu1 (red) and Eu2 (blue) according to STRUCTURE results is shown.