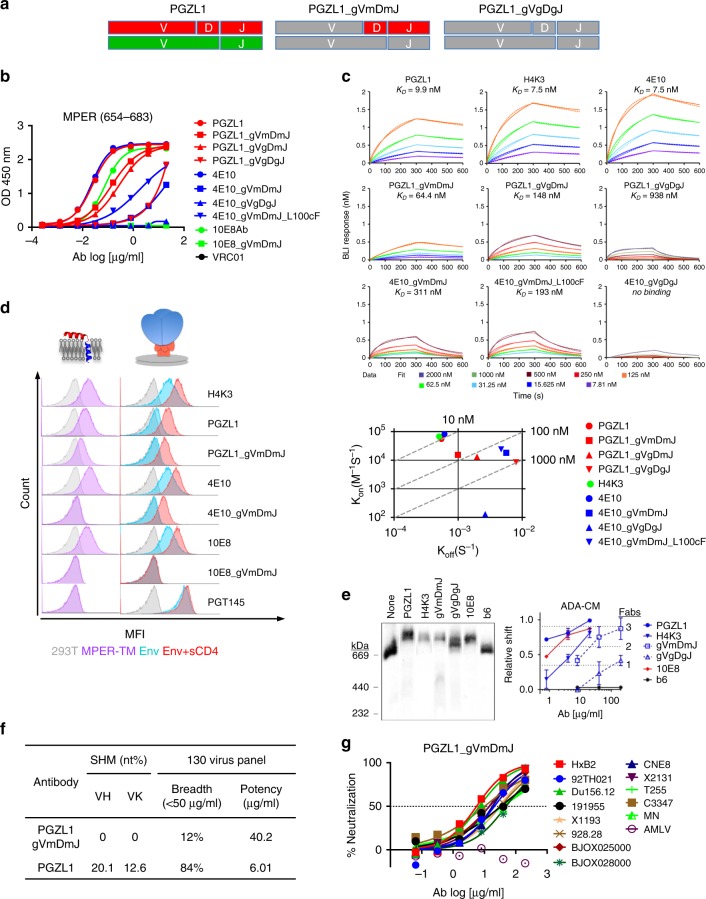
Fig. 2.
Characterization of germline-reverted antibody PGZL1 gVmDmJ. a Cartoon of mature PGZL1 VH (red; top) and Vκ (green; bottom) subdivided by V, D, and J regions, and germline reversions (gray) to create PGZL1 gVmDmJ (middle) and PGZL1 gVgDgJ (right). b ELISA binding of PGZL1 germline revertants to MPER peptide, using analogous 10E8 and 4E10 controls. c BLI-binding kinetics of PGZL1 variants to immobilized MPER peptide (top panels). 4E10 variants were also used for comparison. kon and koff of antibodies are shown on a scatter plot with affinity constant, KD, as dashed lines (bottom). d Cells expressing MPER-TM (purple histograms, left) were stained in flow cytometry by mature and germline-reverted antibodies at 2 μg/ml. HIV Env (right) in the presence and absence of soluble CD4 (sCD4; red and blue histograms, respectively) were stained by mature and germline-reverted antibodies at 2 and 10 μg/ml, respectively. e BN-PAGE Env mobility shift assay. HIV-1 virions were incubated with Fab PGZL1 and H4K3 (20 µg/ml), or PGZL1 gVmDmJ and gVgDgJ (200 µg/ml). 10E8 (20 µg/ml) and non-neutralizing antibody b6 (200 µg/ml) were used as positive and negative controls, respectively. Relative shift and stoichiometry of Fab to Env was quantified. The error bars represent the SD of n = 2 biologically independent experiments. f Neutralization potency and breadth of PGZL1 gVmDmJ against a 130-virus panel of HIV-1 in TZM-bl assay at 200 μg/ml. g Neutralization of 13 isolates sensitive to PGZL1 gVmDmJ chosen from the 130-virus panel. Source data for b–g are provided as a Source Data file.