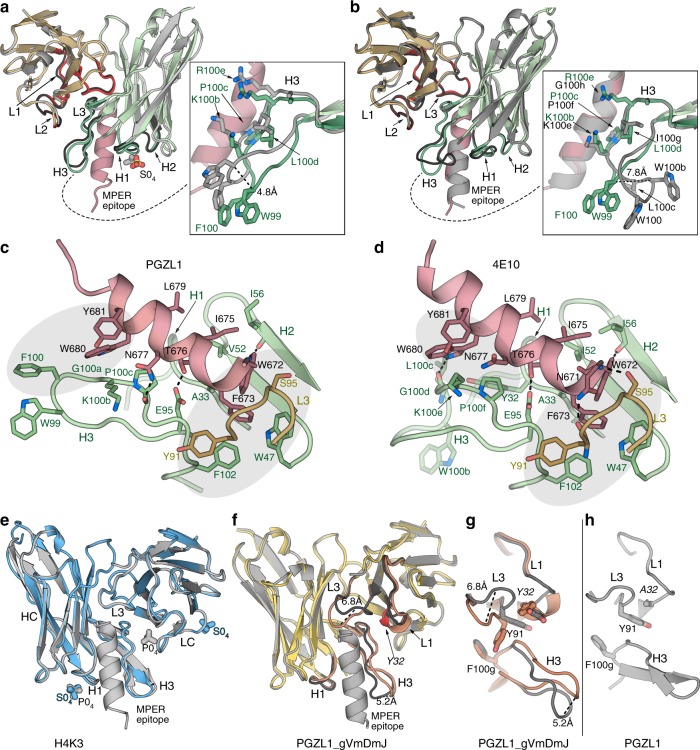
Fig. 4.
PGZL1 variant crystal structures and comparison with 4E10. a Superposition of the crystal structures of the mature PGZL1 variable domain (from the Fab) bound to MPER671-683 (wheat, LC; green, HC; pink, MPER) and unbound PGZL1 (gray). CDRs of the bound structure are shown in red (LC) and green (HC), and CDRs of the unbound structure are shown in black. Inset: superposition of free and bound CDRH3 with residues near the MPER shown as sticks. b Superposition of PGZL1–MPER671-683 and 4E10-MPER671-683 (gray; PDB 2FX7 [https://www.rcsb.org/structure/2fx7]13). Coloring and inset as in a. c PGZL1–MPER671-683 combining site (wheat, LC; green, HC; pink, MPER; interacting residues - sticks). Shaded regions highlight aromatic clusters. d 4E10-MPER671-683 combining site, colored as in c. e Superposition of unbound (blue) and MPER671-683-bound (gray) H4K3. Ions are shown as sticks. f Superposition of unbound (yellow, CDR loops, brown) and MPER671-683-bound (gray) PGZL1 gVmDmJ. g CDR loops in bound (gray) and unbound (brown) PGZL1 gVmDmJ with residues that influence loop conformations shown as sticks. h Same region as in g for the mature PGZL1 structure.