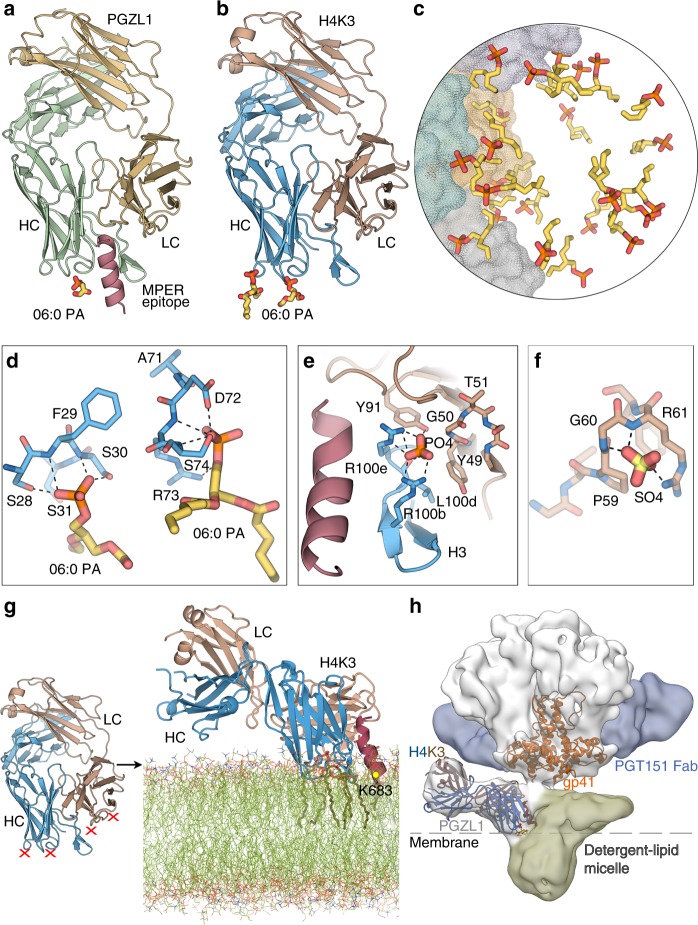
Fig. 5.
Lipid binding and angle of approach of PGZL1 variants to the viral membrane. a Cartoon of the PGZL1-MPER671-683 complex structure (wheat, LC; green, HC; pink, MPER) crystallized with 06:0 PA (sticks). b Cartoon of H4K3 (brown, LC; blue, HC) crystallized with 06:0 PA (sticks). c Stick rendering of 06:0 PA fragments forming a lipid vesicle at the interface of 12 crystallographic and non-crystallographic-related H4K3 Fabs. The four Fabs in the asymmetric unit are shown as gray, green, yellow, and blue color surfaces. d Stick rendering of observed lipid-binding sites in H4K3. e Phosphate-binding site near to H4K3 CDRH3 when MPER671-683 (pink) is bound. Colors as in b. f Sulfate-binding site in FRL3 of H4K3. g Model of H4K3 binding to the MPER (red)-viral membrane (green) epitope. The model at the right side of the arrow was built based on the regions where experimental lipids and anions (red X) bind on H4K3 (left side of the arrow); cognate lipids are shown as sticks inside the modeled membrane. The position of the MPER K683 residue is indicated with a yellow dot. h Cryo-EM reconstruction of full-length AMC011-PGT151-PGZL1 complex at 8.9 Å with H4K3 (blue/brown ribbons) fitted into the Env density at the base of the gp41 stem. The MPER is shown as a red ribbon and lipid head groups as sticks. The detergent–lipid micelle is shown in olive and PGT151 density in blue. Dashed lines show the approximate location where the outer surface of the membrane would be on the virus or infected cells.