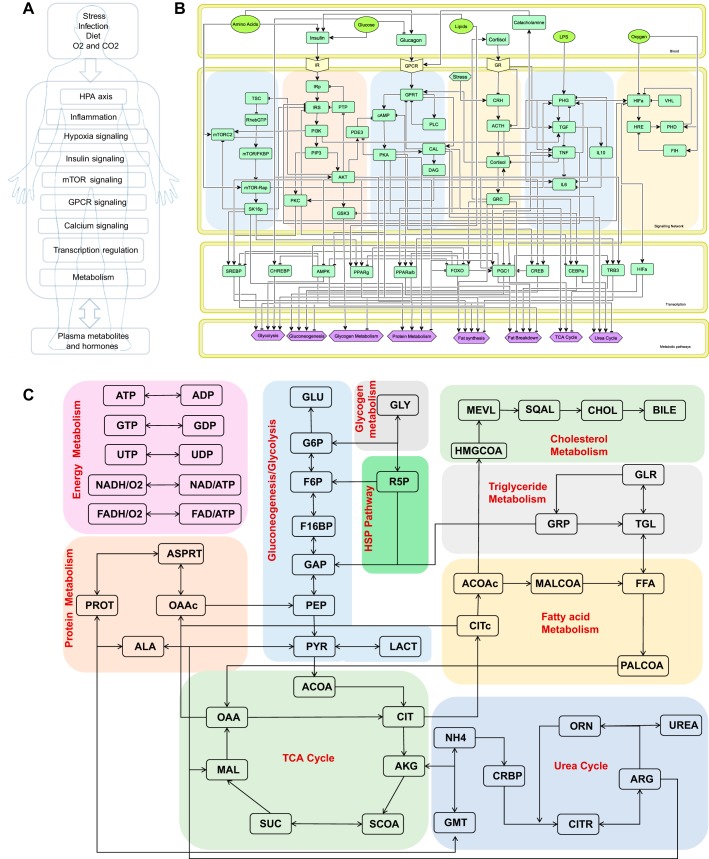
Fig. 3.
A: schematic of the components of the integrated model. B: regulatory network representing 6 signaling pathways, 10 transcription factors, and the metabolic processes that are regulated by the network. Network comprises 1) mammalian target of rapamycin (mTOR) pathway, 2) insulin signaling pathway, 3) G protein-coupled receptor (GPCR) signaling pathway, 4) hypothalamic-pituitary-adrenal (HPA) axis, 5) inflammatory signaling, and 6) hypoxia signaling. These signaling pathways interact to activate downstream transcription regulatory network as shown in the transcription factor compartment. Signaling and transcription network collectively influence metabolic processes as represented in the metabolic pathways compartment. C: metabolic pathways coded in the metabolic simulator. Model constitutes glycolysis, gluconeogenesis, TCA cycle, urea cycle, lipogenesis, amino acid metabolism, hexose amine pathway, pentose phosphate pathway, oxidative phosphorylation, cholesterol pathway, and plasma metabolite fluxes. This model is further integrated with regulations from the regulatory network. ACOAc, acetyl Co-enzyme A (cytosolic); ADP, adenosine diphosphate; AKG, alpha-ketoglutarate; ALA, alanine; ARG, arginine; ASPRT, aspartate; ATP, adenosine triphosphate; CHOL, cholesterol; CIT, citrate; CITc, citrate (cytosolic); CITR, citrulline; CRBP, carbamoyl phosphate; F16BP, fructose-1-6-biphosphate; F6P, fructose-6-phosphate; FAD, flavin adenine dineucleotide; FFA, free fatty acids; GAP, glyceraldehyde phosphate; GDP, guanosine diphosphate; GLR, glycerol; TGL, triglycerides; GLU, glucose; G6P, glucose-6-phosphate; GLY, glycogen; GMT, glutamine; GRP, glycerol-3-phosphate; GTP, guanosine triphosphate; HMGCOA, β-hydroxy-β-methylglutaryl-CoA; LACT, lactate; MAL, malate; MALCOA, malonyl co-enzyme A; MEVL, mevelonate; NAD, nicotinamide adenine dinucleotide; NADP, nicotinamide denine dinucleotide phosphate; NH4, ammonia; OAA, oxaloacetic acid; OAAc, oxaloacetic acid (cytosolic); ORN, ornithine; PALCOA, palmitoyl co-enzyme A; PEP, phopsphoenoyl pyruvate; PROT, protein; PYR, pyruvate; R5P, ribose-5-phosphate; SCOA, succinyl Co-enzyme A; SQAL, squalene; SUC, succinate; UDP, uridine diphosphate; UTP, uridine triphosphate.