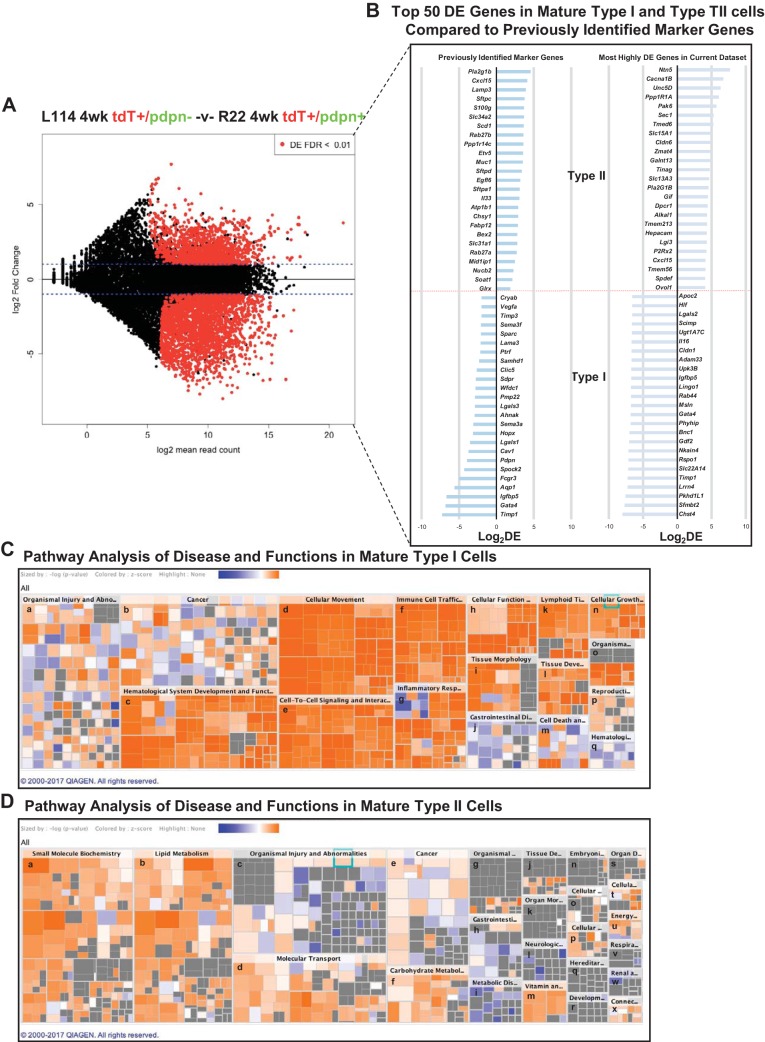
Fig. 6.
Comparison between doxycycline (Dox) E15–18 4 wk TI and TII cells. A: MA plot comparing Dox E15–18 analyzed at 4 wk 114 TdTomato+/Pdpn− (TII cells) and R22 TdTomato+/Pdpn+ (TI cells) shows a large number of DE genes. B: the right column shows the 50 most highly DE protein-encoding genes between PN 4 wk TI and TII cells in the present data set. The left column shows the level of expression, in the present data set, of various previously identified marker genes (7, 17, 31). C and D: analysis of the DE genes by ingenuity pathway analysis reveals substantial differences between the PN 4 wk TI and TII cells. Depicted is a hierarchical heatmap where larger boxes represent a family of related functions and small squares are individual biological processes. The size of the rectangles is correlated with the overlap of predicted and observed gene sets and serves as a significance score (using FET P value). Color indicates the pathways’ predicted activation state (Z score): increasing likelihood (orange) or decreasing likelihood (blue). In in (6C), Tl cell major boxes are associated with genes suggesting newly ascribed Tl cell functions, such as cellular movement, genes relevant to hematological system development, immune cell trafficking, and inflammatory response. In (6D), major boxes of the Tll heatmap are associated with known functions of Tll cells, such as small molecule biochemistry, lipid metabolism, and molecular transport. The image was generated using Qiagen’s IPA software and cropped to reduce the size to improve readability. The unabbreviated names of the pathway names are as follows; in C, PN 4 wk Tl cells: a, organismal injury and abnormalities; b, cancer; c, hematological system development and function; d, cellular movement; e, cell-to-cell signaling and interactions; f, immune cell trafficking; g, inflammatory response; h, cellular function and maintenance; i, tissue morphology; j, gastrointestinal disease; k, lymphoid tissue structure and development; l, tissue development; m, cell death and survival; n, cellular growth and proliferation; o, organismal development; p, reproductive system; q, hematological disease; in D, PN 4 wk Tll cells: a, small molecule biochemistry; b, lipid metabolism; c, organismal injury and abnormalities; d, molecular transport; e, cancer; f, carbohydrate metabolism; g, organismal injury and abnormalities; h, gastrointestinal disease; i, metabolic disease; j, tissue development; k, organ morphology; l, neurological disease; m, vitamin and mineral metabolism; n, embryonic development; o, cellular function and maintenance; p, cellular assembly and organizational; q, heredity disorder; r, developmental disorder; s, organ development; t, cellular assembly and organization; u, energy production; v, respiratory disease; w, renal and urological disease; x, connective tissue development and function. E, embryonic day; PN, postnatal.