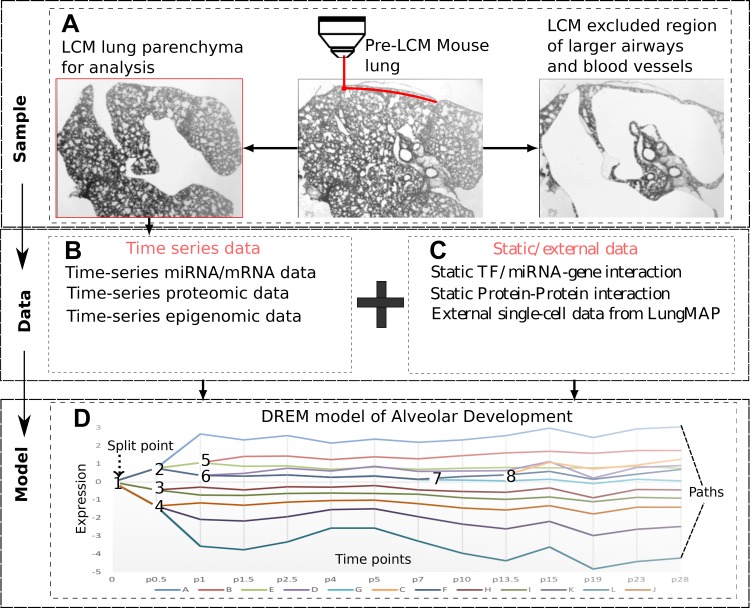
Fig. 1.
Study design. A: samples were obtained from laser capture microdissected (LCM) lungs. B: mRNA, miRNA, and protein levels were profiled at 14 time points selected by the time point selection (TPS) method and methylation at a subset of 6 time points. C: in addition to the condition-specific temporal data the modeling method integrates general static information about protein-DNA and protein-protein interactions and can also use single-cell RNA-Seq (scRNA-Seq) data. D: time-series and static data are integrated using interactive Dynamic Regulatory Events Miner (iDREM). The resulting dynamic model assigns genes to paths and regulators [miRNAs, transcription factors (TFs)] to the paths they regulate. See Fig. 2 for a more detailed version of our model.