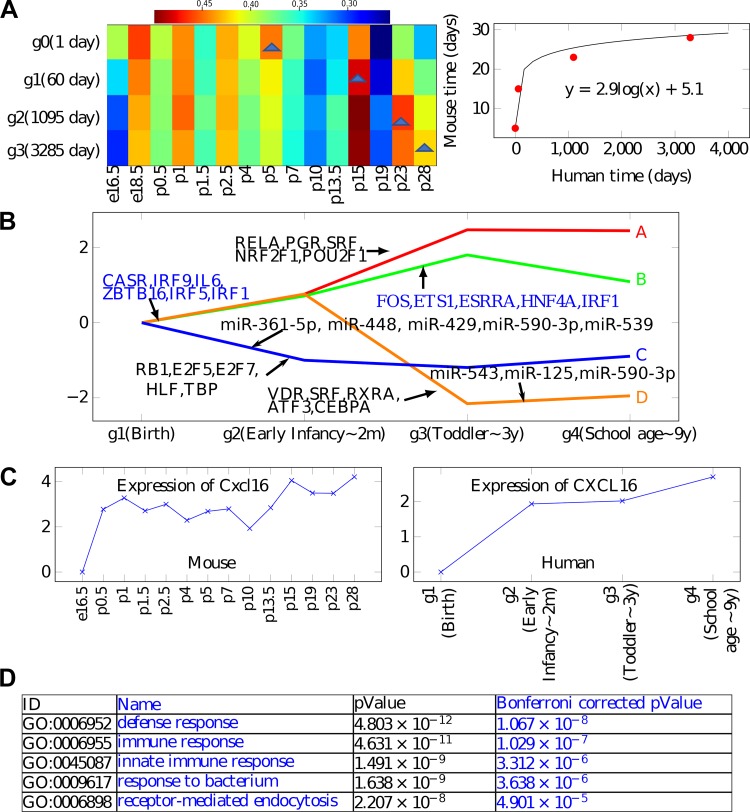
Fig. 4.
Analysis of human data. A: optimal alignment obtained for mouse and human expression data. The color represents the correlation. B: reconstructed human model showing both paths (expression trajectories) and regulators. Note agreement between several of the transcription factors (TFs) and miRNAs in the human and mouse models. C: CXCL16 shows similar expression pattern between the human and mouse data sets and is assigned to a similar path (topmost path) in both models. See Supplemental Fig. S3 (https://doi.org/10.5281/zenodo.3332781) for additional examples of similarly expressed genes. D: Gene Ontology (GO) enrichment for the top path (A) in the human model.