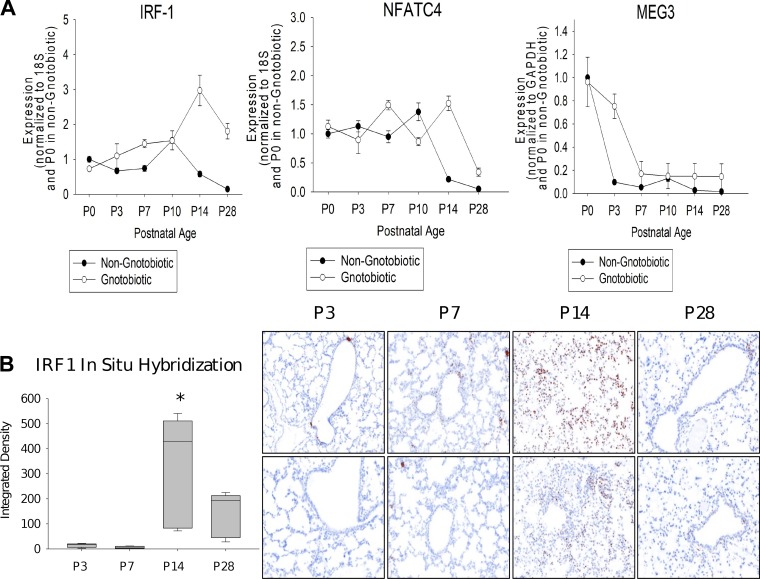
Fig. 5.
Experimental validations of predicted regulators. A: expression of two transcription factors (TFs) and a long non-coding RNA (lncRNA) predicted to regulate key split nodes in the model. As can be seen, even though the model was learned using data from wild-type (WT) mice, we see a comparable increase (for IRF1 and NFATC4) or decrease (Meg3) in the gnotobiotic (germ-free) mice indicating that these regulators are activated or repressed as part of the general developmental program rather than in response to specific pathogens (n = 6 per time point; mean + SE). B: IRF1 localization by in situ hybridization (ISH) in mouse lungs at postnatal day 3 (P3), P7, P14, and P28. Left: image analysis quantitation of IRF1 ISH (n = 3 per time point; box-and-whiskers plot with box indicating 25th–75th percentiles, line across box indicating median, and whiskers indicating 5th–95th percentiles. *P < 0.05 vs. control by 2-way ANOVA followed by Tukey’s test). Right: photomicrographs of ISH demonstrating low concentration of IRF1 at P3 and P7 in a few bronchiolar epithelial and alveolar cells. In contrast, we observe high concentrations at P14 (seen diffusely in many alveolar cells), followed by a decline at P28 with staining only in a few bronchiolar epithelial cells.