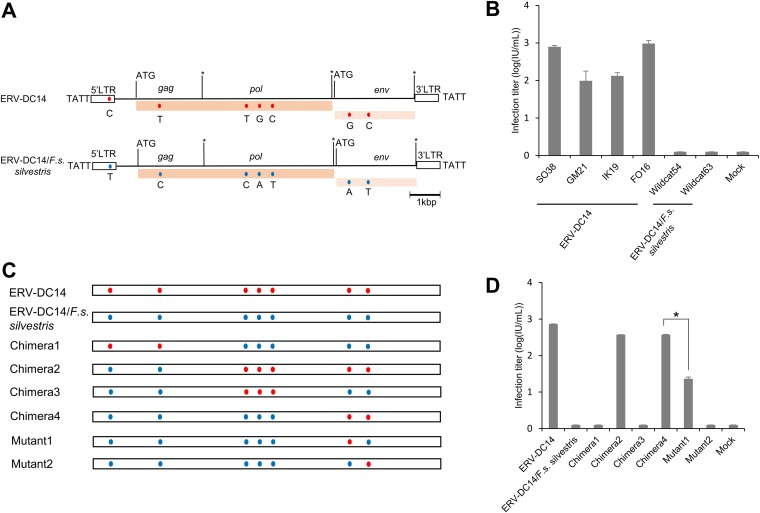
FIG 2.
Characterization and assessment of ERV-DC14/F. s. silvestris. (A) Schematic image of the full-length ERV-DC14/F. s. silvestris provirus. The ERV-DC14 clone SO38 strain was used as the ERV-DC14 reference genome. The gag, pol, and env genes are illustrated together with the 5ʹ and 3ʹ LTRs and the positions of the gag and env translational initiation codons (ATG). Asterisks, stop codons; dark pink box, open reading frame (ORF) of the Gag-Pol polyprotein; light pink box, Env protein; blue and red round circles, single nucleotide polymorphisms (SNPs) between the two proviruses ERV-DC14/SO38 and ERVDC14/F. s. silvestris. Nucleotide substitutions are shown. Flanking 4-bp target duplicate site (TSD) sequences are shown for each provirus. (B) Assessment of the replication-competent activity of ERV-DC14 in European wildcats (F. s. silvestris) and Japanese domestic cats. All tested proviral clones, including ERV-DC14 from different Japanese domestic cats (SO38, GM21, IK19, FO16), ERV-DC14/F. s. silvestris (wildcat 54, and wildcat 63), or the empty vector (mock transfection), were transfected into 293Lac cells, and then their infectivity was tested with fresh HEK293T cells. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations. (C) Schematic representation of the chimeric structures of the two proviruses. Chimera1 to Chimera4 were constructed via recombination between the two proviruses using restriction enzyme digestion, and the two mutants were constructed by site-directed mutagenesis. Blue and red round circles indicate single nucleotide polymorphisms (SNPs) between the two proviruses ERV-DC14/SO38 and ERVDC14/F. s. silvestris. (D) Assessment of the replication competence of chimeric ERV-DC14. 293Lac cells were transfected with plasmids containing different chimeric ERV-DC14s or mock transfected (with the empty vector), and the resulting supernatants were collected and used to infect fresh HEK293T cells. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations. *, P < 0.0001 (one-way ANOVA).