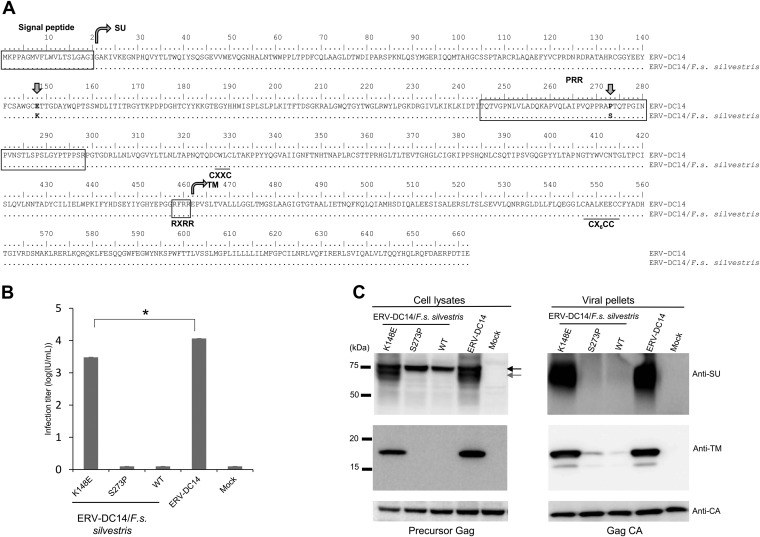
FIG 3.
Determination of the mutation responsible for ERV-DC14/F. s. silvestris Env dysfunction. (A) Amino acid sequence alignment of Env proteins of ERV-DC14 and ERV-DC4/F. s. silvestris. SU, surface subunit; PRR, proline-rich region; TM, transmembrane subunit. RXRR is the cleavage motif. CXXC and CX6CC are sites of covalent interaction. Arrows indicate the positions of amino acids 148 and 273, which differ between these two ERV-DC14 proviruses. (B) Assessment of Env-pseudotyped viruses based on the ERV-DC14/F. s. silvestris wild type (WT), Mutant1 (K148E), and Mutant2 (S273P) or on ERV-DC14. GPLac cells were transfected with the indicated Env expression plasmids. The corresponding Env-pseudotyped viruses were used to infect fresh HEK293T cells. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations. *, P < 0.0001 (one-way ANOVA). (C) Western blotting of GPLac cells expressing ERV-DC14/F. s. silvestris Env (K148E, S273P, or WT) or ERV-DC14 Env. The cell lysates and viral pellets from culture supernatants were analyzed. A goat polyclonal anti-FeLV SU (gp70) antibody was used to detect ERV-DC14 SU, and a mouse monoclonal anti-FeLV TM protein (p15E) antibody was used to detect the ERV-DC14 TM protein. The black arrow indicates immature SU; the gray arrow indicates mature SU. Precursor Gag (Pr65) and Gag CA (p30) were both detected with a goat anti-Raucher MLV CA antibody.