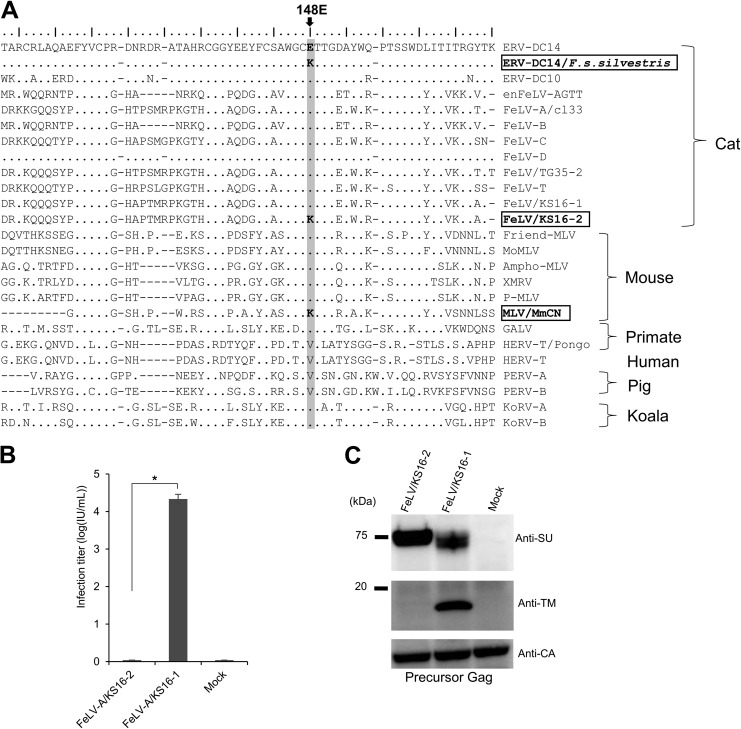
FIG 5.
Sequence analysis of gammaretrovirus Env proteins and dysfunction of a FeLV variant induced by a single mutation in the SU N-terminal domain. (A) Sequence alignment of Env in gammaretroviruses, constructed by use of the mafft tool (70). The amino acid sequences of the gammaretrovirus SU N-terminal regions are also presented. The critical amino acid position 148E is shaded in light gray. The GenBank accession numbers of the reference sequences are as follows: BBL19108.1 for ERV-DC14/F. s. silvestris, BAM33599.1 for ERV-DC14, BAM33597.1 for ERV-DC10, AY364318.1 for enFeLV-AGTT, BAB63924.2 for FeLV-A clone 33 (cl33), AAA43052.1 for FeLV-B, AAA43049.1 for FeLV-C, BAM33588.1 for FeLV-D, BAU61794.1 for FeLV/TG35-2, AAA43050.1 for FeLV-T, BAK41670.2 for FeLV/KS16-1, BBL19109.1 for FeLV/KS16-2, AAA46480.1 for Friend MLV, NP_057935.1 for MoMLV, AAA46515.1 for Ampho-MLV, ADU55755.1 for XMRV, ARB03464.1 for P-MLV, AMK06448.1 for MLV/MmCN, AAA46811.1 for GALV, CAI15393.1 for HERV-T/Pongo, XP_011526770.1 for HERV-T, AAQ83899.1 for PERV-A, BAM67147.1 for KoRV-A, and AGO86848.1 for KoRV-B. (B) Infection assay using pseudotyped viruses of two FeLV variants (KS16-1 and KS16-2). GPLac cells were transfected with Env expression plasmids for FeLV/KS16-1 and FeLV/KS16-2. The filtered viral supernatants were used to infect fresh HEK293T cells. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations. *, P < 0.0001 (one-way ANOVA). (C) Immunoblotting analysis using cell lysates from GPLac cells transfected with the Env expression plasmids which are presented in panel B. Env proteins were detected by use of a mouse anti-FeLV SU (gp70) antibody and an anti-FeLV TM protein (p15E) antibody. Precursor Gag (Pr65) was detected with a goat anti-Raucher MLV CA antibody. The filter exposure time differed for each antibody.