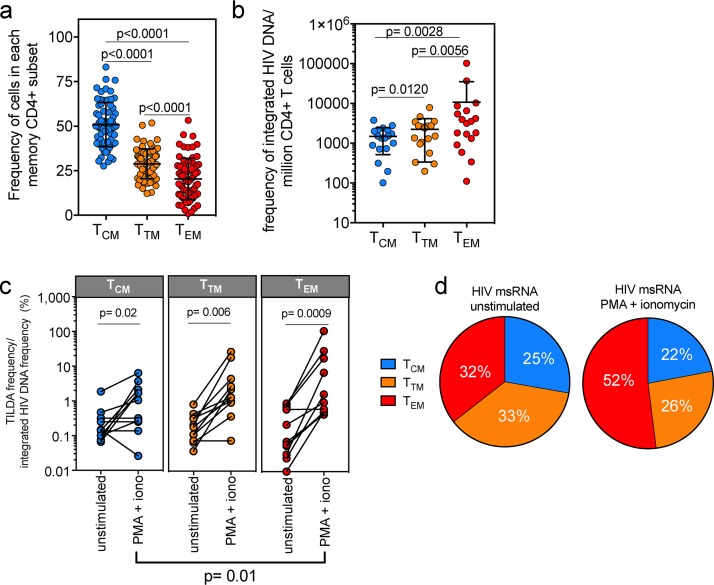
FIG 1.
The inducible HIV reservoir resides in the TEM subset. (a) The distribution of the TCM, TTM, and TEM subsets in ex vivo memory CD4+ T cells is shown, and P values are indicated. Each circle represents individual participants from Table S1 in the supplemental material (bars indicate means with standard deviations [SD], Wilcoxon matched-pair signed-rank test, n = 69). (b) CD4+ T cells from virally suppressed individuals were sorted into TCM, TTM, and TEM subsets using the gating strategy in Fig. S1 in the supplemental material, and integrated HIV DNA in each subset was quantified. Bars indicate means with SD from a Wilcoxon matched-pair signed-rank test; P values are indicated (n = 18). (c) The frequency of transcription of HIV msRNA induced by PMA plus ionomycin (iono), normalized by the amount of integrated HIV DNA in each subset, is shown. Unstimulated sorted subsets were included as a control. There was no statistical difference between levels of HIV msRNA in cells from the CD4+ memory subsets before activation. After exposure to PMA and ionomycin, the frequency of HIV msRNA was quantified and calculated. P values are indicated. (Wilcoxon matched-pair signed-rank test; n = 11). (d) Contribution of HIV msRNA from each CD4+ memory subset from panel c. All undetectable values were input as zero. The contributions of the TCM, TTM, and TEM subsets to the overall HIV msRNA signal are shown. Each pie slice was calculated using the frequency of HIV msRNA in each memory subset as a percentage of the total signal. Frequency is indicated within each pie slice (n = 11).