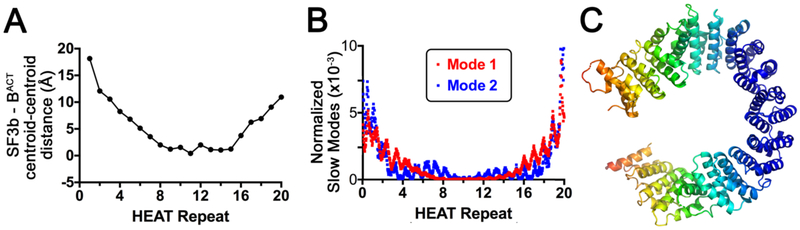
Figure 7.
(A) The distances between the centroids of the HEAT repeats following alignment of the human SF3b1 structures in the SF3b particle (PDB ID: 5IFE) compared to BACT spliceosome (PDB ID: 6FF4). (B-C), Anisotropic network model analysis (ANM 2.0, http://anm.csb.pitt.edu/) shows natural modes of SF3b1 motion similar to its conformational changes for spliceosome integration. The core HEAT domain (residues 463-1274) is analyzed. (B), Plot of the major normalized modes (fluctuations, a measure of flexibility) per SF3b1 residue. The directions of mode 1 (red) and mode 2 (blue) respectively close and twist the SF3b1 torus. (C), Heat map of normalized fluctuations in mode 1, in a color gradient from blue for no movement to orange for maximum movement. Mode 2 appears similar, although the direction of movement differs.