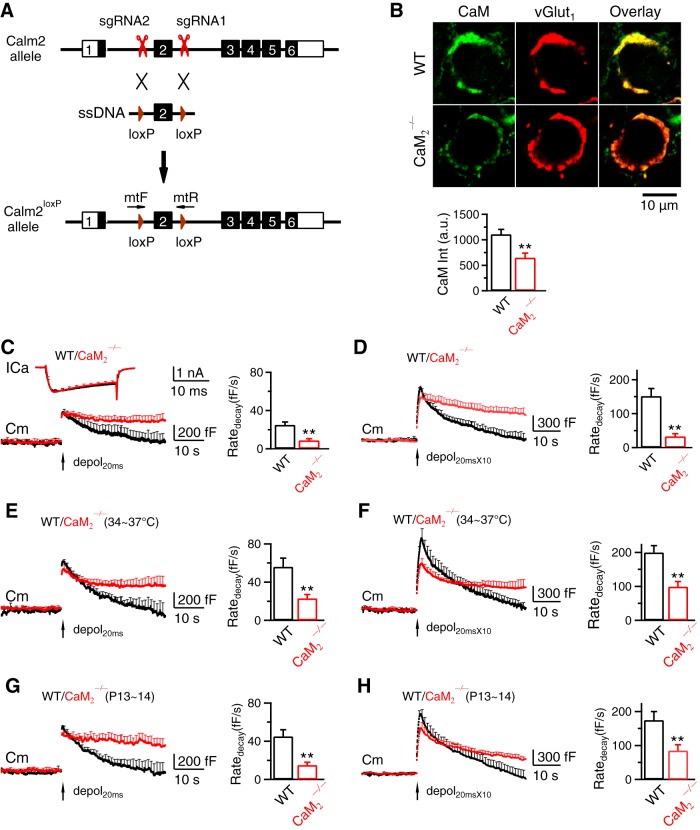
Figure 6.
CaM 2 knock-out inhibits slow endocytosis, rapid endocytosis, and vesicle mobilization to the readily releasable pool at calyces. A, Generation of Calm2loxP mice (Calm2: Calmodulin 2 gene). sgRNAs were designed by using CRISPR Design (https://zlab.bio/guide-design-resources) to identify unique target sites throughout the mouse genome. sgRNAs were transcribed in vitro using the MEGAshortscript T7 Transcription Kit (Life Technologies) from synthetic double-strand DNAs purchased from IDT (Integrated DNA Technologies) and purified using MEGAclear kit (Life Technologies). A mixture of Cas9 mRNA (TriLink Biotechnologies, 100 ng/μl), sgRNAs (50 ng/μl), and ssDNA templates (100 ng/μl, synthesized by IDT) was injected into the cytoplasm of one cell-stage fertilized embryos harvested from C57BL/6J mice (The Jackson Laboratory, 000664). Viable two-cell stage embryos were transferred into the oviducts of female surrogates to generate founder mice. Founders with loxP inserts were identified by PCR and sequencing, and were subsequently bred with C57BL/6J mice to generate heterozygous mice. The primers used to identify the 5′ and 3′ loxP insertions were Calm2 mtF: 5′-CCATGAACCTTGAACCTGTAGGATCCA-3′ and Calm2 mtR: 5′-ATGCTACATTCAACTTGTCACCATTCGAATTCA-3′. B, Top, Antibody staining of CaM and vGluT1 (labeling calyx) in a P9 WT (upper) and a CaM2−/− (lower) calyx (images superimposed in the right). Bottom, CaM staining intensity (mean + SEM, a.u., arbitrary unit) in P7–P10 WT (47 calyces, 4 mice) and CaM2−/− calyces (53 calyces, 4 mice). **p < 0.01 (t test). C, ICa, Cm and Ratedecay (mean + SEM) induced by depol20ms (arrow) in WT (8c/8m, black) and CaM2−/− (9c/9m) calyces (P7–P10, 22–24°C). D, Similar to C, but with depol20msX10 (WT, 8c/8m; CaM2−/−, 9c/9m). E, F, Similar to C and D, respectively, but at 34–37°C. E, WT, 6c/6m; CaM2−/−, 6c/6m. F, WT, 6c/6m; CaM2−/−, 6c/6m. G, H, Similar to C and D, respectively, but from P13–P14 calyces. G, WT, 7c/7m; CaM2−/−, 8c/8m. H, WT, 7c/7m; CaM2−/−, 8c/8m.